Network analysis of transcriptomic diversity amongst resident tissue macrophages and dendritic cells in the mouse mononuclear phagocyte system
- PMID: 33031383
- PMCID: PMC7575120
- DOI: 10.1371/journal.pbio.3000859
Network analysis of transcriptomic diversity amongst resident tissue macrophages and dendritic cells in the mouse mononuclear phagocyte system
Abstract
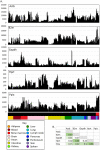
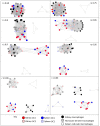
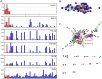
The mononuclear phagocyte system (MPS) is a family of cells including progenitors, circulating blood monocytes, resident tissue macrophages, and dendritic cells (DCs) present in every tissue in the body. To test the relationships between markers and transcriptomic diversity in the MPS, we collected from National Center for Biotechnology Information Gene Expression Omnibus (NCBI-GEO) a total of 466 quality RNA sequencing (RNA-seq) data sets generated from mouse MPS cells isolated from bone marrow, blood, and multiple tissues. The primary data were randomly downsized to a depth of 10 million reads and requantified. The resulting data set was clustered using the network analysis tool BioLayout. A sample-to-sample matrix revealed that MPS populations could be separated based upon tissue of origin. Cells identified as classical DC subsets, cDC1s and cDC2s, and lacking Fcgr1 (encoding the protein CD64) were contained within the MPS cluster, no more distinct than other MPS cells. A gene-to-gene correlation matrix identified large generic coexpression clusters associated with MPS maturation and innate immune function. Smaller coexpression gene clusters, including the transcription factors that drive them, showed higher expression within defined isolated cells, including monocytes, macrophages, and DCs isolated from specific tissues. They include a cluster containing Lyve1 that implies a function in endothelial cell (EC) homeostasis, a cluster of transcripts enriched in intestinal macrophages, and a generic lymphoid tissue cDC cluster associated with Ccr7. However, transcripts encoding Adgre1, Itgax, Itgam, Clec9a, Cd163, Mertk, Mrc1, Retnla, and H2-a/e (encoding class II major histocompatibility complex [MHC] proteins) and many other proposed macrophage subset and DC lineage markers each had idiosyncratic expression profiles. Coexpression of immediate early genes (for example, Egr1, Fos, Dusp1) and inflammatory cytokines and chemokines (tumour necrosis factor [Tnf], Il1b, Ccl3/4) indicated that all tissue disaggregation and separation protocols activate MPS cells. Tissue-specific expression clusters indicated that all cell isolation procedures also co-purify other unrelated cell types that may interact with MPS cells in vivo. Comparative analysis of RNA-seq and single-cell RNA-seq (scRNA-seq) data from the same lung cell populations indicated that MPS heterogeneity implied by global cluster analysis may be even greater at a single-cell level. This analysis highlights the power of large data sets to identify the diversity of MPS cellular phenotypes and the limited predictive value of surface markers to define lineages, functions, or subpopulations.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures






References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

