Mitochondrial Genome Evolution of Placozoans: Gene Rearrangements and Repeat Expansions
- PMID: 33031489
- PMCID: PMC7813641
- DOI: 10.1093/gbe/evaa213
Mitochondrial Genome Evolution of Placozoans: Gene Rearrangements and Repeat Expansions
Abstract
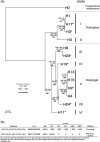
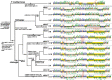
Placozoans, nonbilaterian animals with the simplest known metazoan bauplan, are currently classified into 20 haplotypes belonging to three genera, Polyplacotoma, Trichoplax, and Hoilungia. The latter two comprise two and five clades, respectively. In Trichoplax and Hoilungia, previous studies on six haplotypes belonging to four different clades have shown that their mtDNAs are circular chromosomes of 32-43 kb in size, which encode 12 protein-coding genes, 24 tRNAs, and two rRNAs. These mitochondrial genomes (mitogenomes) also show unique features rarely seen in other metazoans, including open reading frames (ORFs) of unknown function, and group I and II introns. Here, we report seven new mitogenomes, covering the five previously described haplotypes H2, H17, H19, H9, and H11, as well as two new haplotypes, H23 (clade III) and H24 (clade VII). The overall gene content is shared between all placozoan mitochondrial genomes, but genome sizes, gene orders, and several exon-intron boundaries vary among clades. Phylogenomic analyses strongly support a tree topology different from previous 16S rRNA analyses, with clade VI as the sister group to all other Hoilungia clades. We found small inverted repeats in all 13 mitochondrial genomes of the Trichoplax and Hoilungia genera and evaluated their distribution patterns among haplotypes. Because Polyplacotoma mediterranea (H0), the sister to the remaining haplotypes, has a small mitochondrial genome with few small inverted repeats and ORFs, we hypothesized that the proliferation of inverted repeats and ORFs substantially contributed to the observed increase in the size and GC content of the Trichoplax and Hoilungia mitochondrial genomes.
Keywords: Placozoa; evolution; gene order; mitochondrial genome; phylogeny; small inverted repeat.
© The Author(s) 2020. Published by Oxford University Press on behalf of the Society for Molecular Biology and Evolution.
Figures


Similar articles
-
Polyplacotoma mediterranea is a new ramified placozoan species.Curr Biol. 2019 Mar 4;29(5):R148-R149. doi: 10.1016/j.cub.2019.01.068. Curr Biol. 2019. PMID: 30836080
-
Comparative genomics of large mitochondria in placozoans.PLoS Genet. 2007 Jan 12;3(1):e13. doi: 10.1371/journal.pgen.0030013. PLoS Genet. 2007. PMID: 17222063 Free PMC article.
-
Long-Term Culturing of Placozoans (Trichoplax and Hoilungia).Methods Mol Biol. 2024;2757:509-529. doi: 10.1007/978-1-0716-3642-8_21. Methods Mol Biol. 2024. PMID: 38668981
-
The enigmatic Placozoa part 1: Exploring evolutionary controversies and poor ecological knowledge.Bioessays. 2021 Oct;43(10):e2100080. doi: 10.1002/bies.202100080. Epub 2021 Sep 1. Bioessays. 2021. PMID: 34472126 Review.
-
Brief History of Placozoa.Methods Mol Biol. 2024;2757:103-122. doi: 10.1007/978-1-0716-3642-8_3. Methods Mol Biol. 2024. PMID: 38668963 Review.
Cited by
-
Mitogenomics of the zoonotic parasite Echinostoma miyagawai and insights into the evolution of tandem repeat regions within the mitochondrial non-coding control region.Parasitology. 2024 Dec;151(14):1543-1554. doi: 10.1017/S0031182024001422. Parasitology. 2024. PMID: 39540328 Free PMC article.
-
The placozoan Trichoplax.Nat Methods. 2024 Apr;21(4):543-545. doi: 10.1038/s41592-024-02228-3. Nat Methods. 2024. PMID: 38609555 No abstract available.
-
Beauty in the beast - Placozoan biodiversity explored through molluscan predator genomics.Ecol Evol. 2024 Apr 11;14(4):e11220. doi: 10.1002/ece3.11220. eCollection 2024 Apr. Ecol Evol. 2024. PMID: 38606341 Free PMC article.
-
Pervasive Mitochondrial tRNA Gene Loss in Clade B of Haplosclerid Sponges (Porifera, Demospongiae).Genome Biol Evol. 2025 Mar 6;17(3):evaf020. doi: 10.1093/gbe/evaf020. Genome Biol Evol. 2025. PMID: 39913674 Free PMC article.
-
Uncovering the role of mitochondrial genome in pathogenicity and drug resistance in pathogenic fungi.Front Cell Infect Microbiol. 2025 Apr 16;15:1576485. doi: 10.3389/fcimb.2025.1576485. eCollection 2025. Front Cell Infect Microbiol. 2025. PMID: 40308969 Free PMC article. Review.
References
-
- Beck N, Lang B.. 2010. MFannot, organelle genome annotation webserver. Available from: http://megasun.bch.umontreal.ca/cgi-bin/mfannot/mfannotInterface.pl (accessed October 2, 2019).
-
- Burger G, Yan Y, Javadi P, Lang BF.. 2009. Group I-intron trans-splicing and mRNA editing in the mitochondria of placozoan animals. Trends Genet. 25(9):381–386. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

