A Mouse-Adapted SARS-CoV-2 Induces Acute Lung Injury and Mortality in Standard Laboratory Mice
- PMID: 33031744
- PMCID: PMC7510428
- DOI: 10.1016/j.cell.2020.09.050
A Mouse-Adapted SARS-CoV-2 Induces Acute Lung Injury and Mortality in Standard Laboratory Mice
Abstract
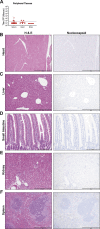
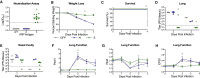
The SARS-CoV-2 pandemic has caused extreme human suffering and economic harm. We generated and characterized a new mouse-adapted SARS-CoV-2 virus that captures multiple aspects of severe COVID-19 disease in standard laboratory mice. This SARS-CoV-2 model exhibits the spectrum of morbidity and mortality of COVID-19 disease as well as aspects of host genetics, age, cellular tropisms, elevated Th1 cytokines, and loss of surfactant expression and pulmonary function linked to pathological features of acute lung injury (ALI) and acute respiratory distress syndrome (ARDS). This model can rapidly access existing mouse resources to elucidate the role of host genetics, underlying molecular mechanisms governing SARS-CoV-2 pathogenesis, and the protective or pathogenic immune responses related to disease severity. The model promises to provide a robust platform for studies of ALI and ARDS to evaluate vaccine and antiviral drug performance, including in the most vulnerable populations (i.e., the aged) using standard laboratory mice.
Keywords: COVID-19; SARS-CoV-2; acute lung injury; acute respiratory distress syndrome; animal models; interferon; mouse-adaptation; vaccines.
Copyright © 2020 Elsevier Inc. All rights reserved.
Conflict of interest statement
Competing Interests The authors declare no competing interests.
Figures




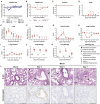


References
-
- Bao L., Deng W., Huang B., Gao H., Liu J., Ren L., Wei Q., Yu P., Xu Y., Qi F., et al. The pathogenicity of SARS-CoV-2 in hACE2 transgenic mice. Nature. 2020;583:830–833. - PubMed
-
- Bolles M., Deming D., Long K., Agnihothram S., Whitmore A., Ferris M., Funkhouser W., Gralinski L., Totura A., Heise M., Baric R.S. A double-inactivated severe acute respiratory syndrome coronavirus vaccine provides incomplete protection in mice and induces increased eosinophilic proinflammatory pulmonary response upon challenge. J. Virol. 2011;85:12201–12215. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

