The mole genome reveals regulatory rearrangements associated with adaptive intersexuality
- PMID: 33033216
- PMCID: PMC8243244
- DOI: 10.1126/science.aaz2582
The mole genome reveals regulatory rearrangements associated with adaptive intersexuality
Abstract
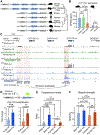
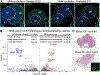
Linking genomic variation to phenotypical traits remains a major challenge in evolutionary genetics. In this study, we use phylogenomic strategies to investigate a distinctive trait among mammals: the development of masculinizing ovotestes in female moles. By combining a chromosome-scale genome assembly of the Iberian mole, Talpa occidentalis, with transcriptomic, epigenetic, and chromatin interaction datasets, we identify rearrangements altering the regulatory landscape of genes with distinct gonadal expression patterns. These include a tandem triplication involving CYP17A1, a gene controlling androgen synthesis, and an intrachromosomal inversion involving the pro-testicular growth factor gene FGF9, which is heterochronically expressed in mole ovotestes. Transgenic mice with a knock-in mole CYP17A1 enhancer or overexpressing FGF9 showed phenotypes recapitulating mole sexual features. Our results highlight how integrative genomic approaches can reveal the phenotypic impact of noncoding sequence changes.
Copyright © 2020 The Authors, some rights reserved; exclusive licensee American Association for the Advancement of Science. No claim to original U.S. Government Works.
Figures



References
-
- Lopez-Rios J et al., Attenuated sensing of SHH by Ptch1 underlies evolution of bovine limbs. Nature. 511, 46–51 (2014). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

