Prostate cancer evolution from multilineage primary to single lineage metastases with implications for liquid biopsy
- PMID: 33033260
- PMCID: PMC7545111
- DOI: 10.1038/s41467-020-18843-5
Prostate cancer evolution from multilineage primary to single lineage metastases with implications for liquid biopsy
Abstract
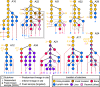
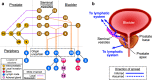
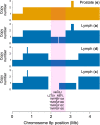
The evolutionary progression from primary to metastatic prostate cancer is largely uncharted, and the implications for liquid biopsy are unexplored. We infer detailed reconstructions of tumor phylogenies in ten prostate cancer patients with fatal disease, and investigate them in conjunction with histopathology and tumor DNA extracted from blood and cerebrospinal fluid. Substantial evolution occurs within the prostate, resulting in branching into multiple spatially intermixed lineages. One dominant lineage emerges that initiates and drives systemic metastasis, where polyclonal seeding between sites is common. Routes to metastasis differ between patients, and likely genetic drivers of metastasis distinguish the metastatic lineage from the lineage that remains confined to the prostate within each patient. Body fluids capture features of the dominant lineage, and subclonal expansions that occur in the metastatic phase are non-uniformly represented. Cerebrospinal fluid analysis reveals lineages not detected in blood-borne DNA, suggesting possible clinical utility.
Conflict of interest statement
All authors delare no competing interests.
Figures



References
-
- Siegel, R. L., Miller, K. D. & Jemal, A. Cancer statistics, 2019. CA. Cancer J. Clin. 10.3322/caac.21551. (2019). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

