Lipocalin-type prostaglandin D synthase regulates light-induced phase advance of the central circadian rhythm in mice
- PMID: 33033338
- PMCID: PMC7544906
- DOI: 10.1038/s42003-020-01281-w
Lipocalin-type prostaglandin D synthase regulates light-induced phase advance of the central circadian rhythm in mice
Abstract
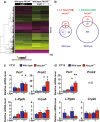
We previously showed that mice lacking pituitary adenylate cyclase-activating polypeptide (PACAP) exhibit attenuated light-induced phase shift. To explore the underlying mechanisms, we performed gene expression analysis of laser capture microdissected suprachiasmatic nuclei (SCNs) and found that lipocalin-type prostaglandin (PG) D synthase (L-PGDS) is involved in the impaired response to light stimulation in the late subjective night in PACAP-deficient mice. L-PGDS-deficient mice also showed impaired light-induced phase advance, but normal phase delay and nonvisual light responses. Then, we examined the receptors involved in the response and observed that mice deficient for type 2 PGD2 receptor DP2/CRTH2 (chemoattractant receptor homologous molecule expressed on Th2 cells) show impaired light-induced phase advance. Concordant results were observed using the selective DP2/CRTH2 antagonist CAY10471. These results indicate that L-PGDS is involved in a mechanism of light-induced phase advance via DP2/CRTH2 signaling.
Conflict of interest statement
The authors declare no competing interests.
Figures




References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

