Potential zoonotic sources of SARS-CoV-2 infections
- PMID: 33034151
- PMCID: PMC7675418
- DOI: 10.1111/tbed.13872
Potential zoonotic sources of SARS-CoV-2 infections
Abstract
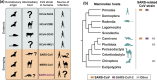
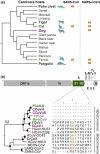
The severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) causing coronavirus disease-2019 (COVID-19) likely has evolutionary origins in other animals than humans based on genetically related viruses existing in rhinolophid bats and pangolins. Similar to other animal coronaviruses, SARS-CoV-2 contains a functional furin cleavage site in its spike protein, which may broaden the SARS-CoV-2 host range and affect pathogenesis. Whether ongoing zoonotic infections are possible in addition to efficient human-to-human transmission remains unclear. In contrast, human-to-animal transmission can occur based on evidence provided from natural and experimental settings. Carnivores, including domestic cats, ferrets and minks, appear to be particularly susceptible to SARS-CoV-2 in contrast to poultry and other animals reared as livestock such as cattle and swine. Epidemiologic evidence supported by genomic sequencing corroborated mink-to-human transmission events in farm settings. Airborne transmission of SARS-CoV-2 between experimentally infected cats additionally substantiates the possibility of cat-to-human transmission. To evaluate the COVID-19 risk represented by domestic and farmed carnivores, experimental assessments should include surveillance and health assessment of domestic and farmed carnivores, characterization of the immune interplay between SARS-CoV-2 and carnivore coronaviruses, determination of the SARS-CoV-2 host range beyond carnivores and identification of human risk groups such as veterinarians and farm workers. Strategies to mitigate the risk of zoonotic SARS-CoV-2 infections may have to be developed in a One Health framework and non-pharmaceutical interventions may have to consider free-roaming animals and the animal farming industry.
Keywords: COVID-19; SARS-CoV-2; carnivore; coronavirus; domestic animal; farmed animal.
© 2020 The Authors. Transboundary and Emerging Diseases published by Wiley-VCH GmbH.
Conflict of interest statement
Authors declare no competing interests.
Figures

References
-
- Alharbi, N. K. , Qasim, I. , Almasoud, A. , Aljami, H. A. , Alenazi, M. W. , Alhafufi, A. , … Balkhy, H. H. (2019). Humoral immunogenicity and efficacy of a single dose of ChAdOx1 MERS vaccine candidate in dromedary camels. Scientific Reports, 9(1), 16292. 10.1038/s41598-019-52730-4. - DOI - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

