DockCoV2: a drug database against SARS-CoV-2
- PMID: 33035337
- PMCID: PMC7778986
- DOI: 10.1093/nar/gkaa861
DockCoV2: a drug database against SARS-CoV-2
Abstract
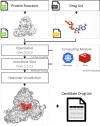
The current state of the COVID-19 pandemic is a global health crisis. To fight the novel coronavirus, one of the best-known ways is to block enzymes essential for virus replication. Currently, we know that the SARS-CoV-2 virus encodes about 29 proteins such as spike protein, 3C-like protease (3CLpro), RNA-dependent RNA polymerase (RdRp), Papain-like protease (PLpro), and nucleocapsid (N) protein. SARS-CoV-2 uses human angiotensin-converting enzyme 2 (ACE2) for viral entry and transmembrane serine protease family member II (TMPRSS2) for spike protein priming. Thus in order to speed up the discovery of potential drugs, we develop DockCoV2, a drug database for SARS-CoV-2. DockCoV2 focuses on predicting the binding affinity of FDA-approved and Taiwan National Health Insurance (NHI) drugs with the seven proteins mentioned above. This database contains a total of 3,109 drugs. DockCoV2 is easy to use and search against, is well cross-linked to external databases, and provides the state-of-the-art prediction results in one site. Users can download their drug-protein docking data of interest and examine additional drug-related information on DockCoV2. Furthermore, DockCoV2 provides experimental information to help users understand which drugs have already been reported to be effective against MERS or SARS-CoV. DockCoV2 is available at https://covirus.cc/drugs/.
© The Author(s) 2020. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

