Genome-Wide Identification of Host-Segregating Single-Nucleotide Polymorphisms for Source Attribution of Clinical Campylobacter coli Isolates
- PMID: 33036986
- PMCID: PMC7688228
- DOI: 10.1128/AEM.01787-20
Genome-Wide Identification of Host-Segregating Single-Nucleotide Polymorphisms for Source Attribution of Clinical Campylobacter coli Isolates
Abstract
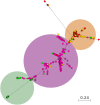
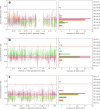
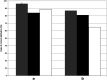
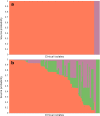
Campylobacter is among the most common causes of gastroenteritis worldwide. Campylobacter jejuni and Campylobacter coli are the most common species causing human disease. DNA sequence-based methods for strain characterization have focused largely on C. jejuni, responsible for 80 to 90% of infections, meaning that C. coli epidemiology has lagged behind. Here, we have analyzed the genome of 450 C. coli isolates to determine genetic markers that can discriminate isolates sampled from 3 major reservoir hosts (chickens, cattle, and pigs). These markers then were applied to identify the source of infection of 147 C. coli strains from French clinical cases. Using STRUCTURE software, 259 potential host-segregating markers were revealed by probabilistic characterization of single-nucleotide polymorphism (SNP) frequency variation in strain collections from three different hosts. These SNPs were found in 41 genes or intergenic regions, mostly coding for proteins involved in motility and membrane functions. Source attribution of clinical isolates based on the differential presence of these markers confirmed chickens as the most common source of C. coli infection in France.IMPORTANCE Genome-wide and source attribution studies based on Campylobacter species have shown their importance for the understanding of foodborne infections. Although the use of multilocus sequence typing based on 7 genes from C. jejuni is a powerful method to structure populations, when applied to C. coli, results have not clearly demonstrated its robustness. Therefore, we aim to provide more accurate data based on the identification of single-nucleotide polymorphisms. Results from this study reveal an important number of host-segregating SNPs, found in proteins involved in motility, membrane functions, or DNA repair systems. These findings offer new, interesting opportunities for further study of C. coli adaptation to its environment. Additionally, the results demonstrate that poultry is potentially the main reservoir of C. coli in France.
Keywords: Campylobacter coli; SNP; genomics; genotyping; source attribution.
Copyright © 2020 American Society for Microbiology.
Figures

References
-
- Van Cauteren D, Le Strat Y, Sommen C, Bruyand M, Tourdjman M, Da Silva NJ, Couturier E, Fournet N, de Valk H, Desenclos J-C. 2017. Estimated annual numbers of foodborne pathogen-associated illnesses, hospitalizations, and deaths, France, 2008–2013. Emerg Infect Dis 23:1486–1492. doi:10.3201/eid2309.170081. - DOI - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical

