Deconvolution of monocyte responses in inflammatory bowel disease reveals an IL-1 cytokine network that regulates IL-23 in genetic and acquired IL-10 resistance
- PMID: 33037057
- PMCID: PMC8108288
- DOI: 10.1136/gutjnl-2020-321731
Deconvolution of monocyte responses in inflammatory bowel disease reveals an IL-1 cytokine network that regulates IL-23 in genetic and acquired IL-10 resistance
Abstract
Objective: Dysregulated immune responses are the cause of IBDs. Studies in mice and humans suggest a central role of interleukin (IL)-23-producing mononuclear phagocytes in disease pathogenesis. Mechanistic insights into the regulation of IL-23 are prerequisite for selective IL-23 targeting therapies as part of personalised medicine.
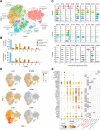
Design: We performed transcriptomic analysis to investigate IL-23 expression in human mononuclear phagocytes and peripheral blood mononuclear cells. We investigated the regulation of IL-23 expression and used single-cell RNA sequencing to derive a transcriptomic signature of hyperinflammatory monocytes. Using gene network correlation analysis, we deconvolved this signature into components associated with homeostasis and inflammation in patient biopsy samples.
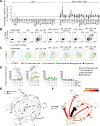
Results: We characterised monocyte subsets of healthy individuals and patients with IBD that express IL-23. We identified autosensing and paracrine sensing of IL-1α/IL-1β and IL-10 as key cytokines that control IL-23-producing monocytes. Whereas Mendelian genetic defects in IL-10 receptor signalling induced IL-23 secretion after lipopolysaccharide stimulation, whole bacteria exposure induced IL-23 production in controls via acquired IL-10 signalling resistance. We found a transcriptional signature of IL-23-producing inflammatory monocytes that predicted both disease and resistance to antitumour necrosis factor (TNF) therapy and differentiated that from an IL-23-associated lymphocyte differentiation signature that was present in homeostasis and in disease.
Conclusion: Our work identifies IL-10 and IL-1 as critical regulators of monocyte IL-23 production. We differentiate homeostatic IL-23 production from hyperinflammation-associated IL-23 production in patients with severe ulcerating active Crohn's disease and anti-TNF treatment non-responsiveness. Altogether, we identify subgroups of patients with IBD that might benefit from IL-23p19 and/or IL-1α/IL-1β-targeting therapies upstream of IL-23.
Keywords: inflammatory bowel disease; interleukins; mucosal immunology.
© Author(s) (or their employer(s)) 2021. Re-use permitted under CC BY. Published by BMJ.
Conflict of interest statement
Competing interests: BS, SH, KC and JS are current or previous employees of Eli Lilly. HU received research support or consultancy fees from UCB Pharma, Eli Lilly, Boehringer Ingelheim, Pfizer, Celgene, OMass and AbVie. FP has received research support or consultancy fees from GSK, UCB Pharma, Medimmune, Janssen and Eli Lilly. SPLT has been adviser to, in receipt of educational or research grants from, or invited lecturer for AbbVie, Amgen, Asahi, Biogen, Boehringer Ingelheim, BMS, Cosmo, Elan, Enterome, Ferring, FPRT Bio, Genentech/Roche, Genzyme, Glenmark, GW Pharmaceuticals, Janssen, Johnson & Johnson, Eli Lilly, Merck, Novartis, Novo Nordisk, Ocera, Pfizer, Shire, Santarus, SigmoidPharma, Synthon, Takeda, Tillotts, Topivert, Trino Therapeutics with Wellcome Trust, UCB Pharma, Vertex, VHsquared, Vifor, Warner Chilcott and Zeria. SK has received consultancy fees from Janssen and Takeda.
Figures





References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
