Single-cell and spatial transcriptomics enables probabilistic inference of cell type topography
- PMID: 33037292
- PMCID: PMC7547664
- DOI: 10.1038/s42003-020-01247-y
Single-cell and spatial transcriptomics enables probabilistic inference of cell type topography
Abstract
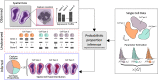
The field of spatial transcriptomics is rapidly expanding, and with it the repertoire of available technologies. However, several of the transcriptome-wide spatial assays do not operate on a single cell level, but rather produce data comprised of contributions from a - potentially heterogeneous - mixture of cells. Still, these techniques are attractive to use when examining complex tissue specimens with diverse cell populations, where complete expression profiles are required to properly capture their richness. Motivated by an interest to put gene expression into context and delineate the spatial arrangement of cell types within a tissue, we here present a model-based probabilistic method that uses single cell data to deconvolve the cell mixtures in spatial data. To illustrate the capacity of our method, we use data from different experimental platforms and spatially map cell types from the mouse brain and developmental heart, which arrange as expected.
Conflict of interest statement
The authors declare no competing interests.
Figures


References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

