Blood RNA signatures predict recent tuberculosis exposure in mice, macaques and humans
- PMID: 33037303
- PMCID: PMC7547102
- DOI: 10.1038/s41598-020-73942-z
Blood RNA signatures predict recent tuberculosis exposure in mice, macaques and humans
Abstract
Tuberculosis (TB) is the leading cause of death due to a single infectious disease. Knowing when a person was infected with Mycobacterium tuberculosis (M.tb) is critical as recent infection is the strongest clinical risk factor for progression to TB disease in immunocompetent individuals. However, time since M.tb infection is challenging to determine in routine clinical practice. To define a biomarker for recent TB exposure, we determined whether gene expression patterns in blood RNA correlated with time since M.tb infection or exposure. First, we found RNA signatures that accurately discriminated early and late time periods after experimental infection in mice and cynomolgus macaques. Next, we found a 6-gene blood RNA signature that identified recently exposed individuals in two independent human cohorts, including adult household contacts of TB cases and adolescents who recently acquired M.tb infection. Our work supports the need for future longitudinal studies of recent TB contacts to determine whether biomarkers of recent infection can provide prognostic information of TB disease risk in individuals and help map recent transmission in communities.
Conflict of interest statement
R.C.A. is inventor on pending patents filed by Texas Biomedical Research Institute for using RNA expression to determine duration of mycobacterial infection, US provisional Patent No. 62/768,708, PCT Patent Application No. PCT/US19/61895. All other authors have no competing interests.
Figures

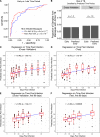
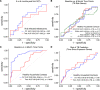

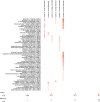
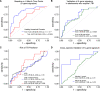
References
-
- World Health Organization. Global Tuberculosis Report 2018. https://www.apps.who.int/medicinedocs/en/m/abstract/Js23553en/.
-
- World Health Organization. Global strategy and targets for tuberculosis prevention, care and control after 2015. https://www.who.int/tb/post2015_strategy/en/ (2014).
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

