A compromised specific humoral immune response against the SARS-CoV-2 receptor-binding domain is related to viral persistence and periodic shedding in the gastrointestinal tract
- PMID: 33037400
- PMCID: PMC7546387
- DOI: 10.1038/s41423-020-00550-2
A compromised specific humoral immune response against the SARS-CoV-2 receptor-binding domain is related to viral persistence and periodic shedding in the gastrointestinal tract
Abstract
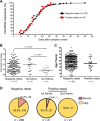
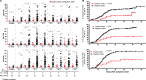
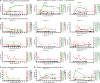
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has been redetected after discharge in some coronavirus disease 2019 (COVID-19) patients. The reason for the recurrent positivity of the test and the potential public health concern due to this occurrence are still unknown. Here, we analyzed the viral data and clinical manifestations of 289 domestic Chinese COVID-19 patients and found that 21 individuals (7.3%) were readmitted for hospitalization after detection of SARS-CoV-2 after discharge. First, we experimentally confirmed that the virus was involved in the initial infection and was not a secondary infection. In positive retests, the virus was usually found in anal samples (15 of 21, 71.4%). Through analysis of the intracellular viral subgenomic messenger RNA (sgmRNA), we verified that positive retest patients had active viral replication in their gastrointestinal tracts (3 of 16 patients, 18.7%) but not in their respiratory tracts. Then, we found that viral persistence was not associated with high viral titers, delayed viral clearance, old age, or more severe clinical symptoms during the first hospitalization. In contrast, viral rebound was associated with significantly lower levels of and slower generation of viral receptor-binding domain (RBD)-specific IgA and IgG antibodies. Our study demonstrated that the positive retest patients failed to create a robust protective humoral immune response, which might result in SARS-CoV-2 persistence in the gastrointestinal tract and possibly in active viral shedding. Further exploration of the mechanism underlying the rebound in SARS-CoV-2 in this population will be crucial for preventing virus spread and developing effective vaccines.
Keywords: SARS-CoV-2; gastrointestinal infection; protective antibody; virus recurrence.
Conflict of interest statement
The authors declare no competing interests.
Figures

References
-
- Hao, X. et al. Reconstruction of the full transmission dynamics of COVID-19 in Wuhan. Nature. 10.1038/s41586-020-2554-8 (2020). - PubMed
-
- He X, et al. Temporal dynamics in viral shedding and transmissibility of COVID-19. Nat. Med. 2020;26:672–675. - PubMed
-
- Wu, Z. & McGoogan, J. M. Characteristics of and important lessons from the Coronavirus Disease 2019 (COVID-19) Outbreak in China: summary of a Report of 72314 Cases From the Chinese Center for Disease Control and Prevention. J. Am. Med. Assoc.10.1001/jama.2020.2648 (2020). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

