The Bgee suite: integrated curated expression atlas and comparative transcriptomics in animals
- PMID: 33037820
- PMCID: PMC7778977
- DOI: 10.1093/nar/gkaa793
The Bgee suite: integrated curated expression atlas and comparative transcriptomics in animals
Abstract
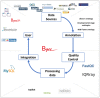
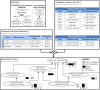
Bgee is a database to retrieve and compare gene expression patterns in multiple animal species, produced by integrating multiple data types (RNA-Seq, Affymetrix, in situ hybridization, and EST data). It is based exclusively on curated healthy wild-type expression data (e.g., no gene knock-out, no treatment, no disease), to provide a comparable reference of normal gene expression. Curation includes very large datasets such as GTEx (re-annotation of samples as 'healthy' or not) as well as many small ones. Data are integrated and made comparable between species thanks to consistent data annotation and processing, and to calls of presence/absence of expression, along with expression scores. As a result, Bgee is capable of detecting the conditions of expression of any single gene, accommodating any data type and species. Bgee provides several tools for analyses, allowing, e.g., automated comparisons of gene expression patterns within and between species, retrieval of the prefered conditions of expression of any gene, or enrichment analyses of conditions with expression of sets of genes. Bgee release 14.1 includes 29 animal species, and is available at https://bgee.org/ and through its Bioconductor R package BgeeDB.
© The Author(s) 2020. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures

References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

