COVID-19: Discovery, diagnostics and drug development
- PMID: 33038433
- PMCID: PMC7543767
- DOI: 10.1016/j.jhep.2020.09.031
COVID-19: Discovery, diagnostics and drug development
Abstract
Coronavirus disease 2019 (COVID-19) started as an epidemic in Wuhan in 2019, and has since become a pandemic. Groups from China identified and sequenced the virus responsible for COVID-19, named severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), and determined that it was a novel coronavirus sharing high sequence identity with bat- and pangolin-derived SARS-like coronaviruses, suggesting a zoonotic origin. SARS-CoV-2 is a member of the Coronaviridae family of enveloped, positive-sense, single-stranded RNA viruses that infect a broad range of vertebrates. The rapid release of the sequence of the virus has enabled the development of diagnostic tools. Additionally, serological tests can now identify individuals who have been infected. SARS-CoV-2 infection is associated with a fatality rate of around 1-3%, which is commonly linked to the development of acute respiratory distress syndrome (ARDS), likely resulting from uncontrolled immune activation, the so called "cytokine storm". Risk factors for mortality include advanced age, obesity, diabetes, and hypertension. Drug repurposing has been used to rapidly identify potential treatments for COVID-19, which could move quickly to phase III. Better knowledge of the virus and its enzymes will aid the development of more potent and specific direct-acting antivirals. In the long term, a vaccine to prevent infection is crucial; however, even if successful, it might not be available before 2021-22. To date, except for intravenous remdesivir and dexamethasone, which have modest effects in moderate to severe COVID-19, no strong clinical evidence supports the efficacy of any other drugs against SARS-CoV-2. The aim of this review is to provide insights on the discovery of SARS-CoV-2, its virology, diagnostic tools, and the ongoing drug discovery effort.
Keywords: Coronavirus; Drug repurposing; Pathogenesis; Remdesivir; SARS-CoV-2.
Copyright © 2020 European Association for the Study of the Liver. All rights reserved.
Conflict of interest statement
Conflict of interest Tarik Asselah has acted as a speaker and investigator for AbbVie, Janssen, Gilead, Roche, and Merck. David Durantel, Eric Pasmant and George Lau have nothing to declare. Raymond Schinazi was an unpaid consultant for Lilly and holds equity in Lilly and Gilead. Please refer to the accompanying ICMJE disclosure forms for further details.
Figures

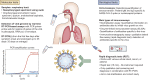



References
-
- WHO Covid-19 situation report 181. Available at: https://www.who.int/. Accessed 4 September 2020.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

