APOBEC: A molecular driver in cervical cancer pathogenesis
- PMID: 33038491
- PMCID: PMC7539941
- DOI: 10.1016/j.canlet.2020.10.004
APOBEC: A molecular driver in cervical cancer pathogenesis
Abstract
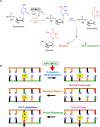
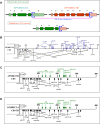
Cervical cancer is one of the foremost common cancers in women. Human papillomavirus (HPV) infection remains a major risk factor of cervical cancer. In addition, numerous other genetic and epigenetic factors also are involved in the underlying pathogenesis of cervical cancer. Recently, it has been reported that apolipoprotein B mRNA editing enzyme catalytic polypeptide like (APOBEC), DNA-editing protein plays an important role in the molecular pathogenesis of cancer. Particularly, the APOBEC3 family was shown to induce tumor mutations by aberrant DNA editing mechanism. In general, APOBEC3 enzymes play a pivotal role in the deamination of cytidine to uridine in DNA and RNA to control diverse biological processes such as regulation of protein expression, innate immunity, and embryonic development. Innate antiviral activity of the APOBEC3 family members restrict retroviruses, endogenous retro-element, and DNA viruses including the HPV that is the leading risk factor for cervical cancer. This review briefly describes the pathogenesis of cervical cancer and discusses in detail the recent findings on the role of APOBEC in the molecular pathogenesis of cervical cancer.
Keywords: Cytidine deaminase; DNA/RNA editing; HPV; Mutation; Viral restriction.
Copyright © 2020 Elsevier B.V. All rights reserved.
Conflict of interest statement
The authors have no conflict of interest to declare.
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

