Immunoinformatics approach to understand molecular interaction between multi-epitopic regions of SARS-CoV-2 spike-protein with TLR4/MD-2 complex
- PMID: 33039603
- PMCID: PMC7543713
- DOI: 10.1016/j.meegid.2020.104587
Immunoinformatics approach to understand molecular interaction between multi-epitopic regions of SARS-CoV-2 spike-protein with TLR4/MD-2 complex
Abstract
Background: The coronavirus (CoV) spike (S) protein is critical for receptor binding, membrane fusion and internalization of the virus into the human cells. We have tried to search the epitopic component of the S-protein that might be served as crucial targets for the vaccine development and also tried to understand the molecular mechanism of epitopes and TLR4/MD-2 complex for adaptive immunity.
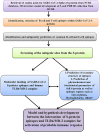
Material and methods: Here we identified the antigenicity and the epitopic divergence of S-protein via immunoinformatics approach. The study was performed to identify the epitopes, composition of amino acids and its distribution in epitopic regions, composition of amino acid between the identified epitopes, secondary structure architecture of epitopes, physicochemical and biochemical parameters and molecular interaction between the identified epitope and TLR4/MD-2 complex. The SARS-CoV-2 can be possibly recognised by TLR4 of host immune cells that are responsible for the adaptive immune response.
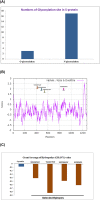
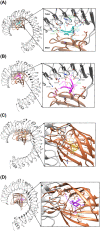
Results: We identified four SARS-CoV-2 S-protein 9mer antigenic epitopes and observed that they bind with the TLR4/MD-2 complex by varied stable molecular bonding interactions. Molecular interaction between these characterized epitopes with TLR4/MD-2 complex might be indicated the binding affinity and downstream signalling of adaptive immune response. Different physicochemical and biochemical parameters such as O-glycosylation and N-glycosylation, Hydrophobicity, GRAVY were identified within epitopic regions of S-protein. These parameters help to understand the protein-protein interaction between epitopes and TLR4/MD-2 complex. The study also revealed different epitopic binding pockets of TLR4/MD-2 complex.
Conclusions: The identified epitopes impart suitable prospects for the development of novel peptide-based epitopic vaccine for the control of COVID-19 infection.
Keywords: Antigenicity; Epitopes; SARS-CoV-2; Spike protein; TLR4/MD-2 complex.
Copyright © 2020 Elsevier B.V. All rights reserved.
Conflict of interest statement
All of the authors declare that there is no competing interest in this work.
Figures





References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

