USP7 mediates pathological hepatic de novo lipogenesis through promoting stabilization and transcription of ZNF638
- PMID: 33040080
- PMCID: PMC7548010
- DOI: 10.1038/s41419-020-03075-8
USP7 mediates pathological hepatic de novo lipogenesis through promoting stabilization and transcription of ZNF638
Abstract
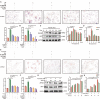
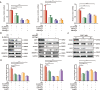
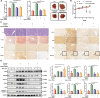
Aberrant de novo lipogenesis (DNL) results in excessive hepatic lipid accumulation and liver steatosis, the causative factors of many liver diseases, such as non-alcoholic fatty liver disease (NAFLD), non-alcoholic steatohepatitis (NASH), and hepatocellular carcinoma (HCC). However, the underlying mechanism of DNL dysregulation remains largely unknown. Ubiquitination of proteins in hepatocytes has been shown to be widely involved in lipid metabolism of liver. Here, we revealed that Ubiquitin-specific peptidase 7 (USP7), a deubiquitinase (DUB), played key roles in DNL through regulation of zinc finger protein 638 (ZNF638) in hepatocytes. USP7 has been shown not only to interact with and deubiquitylate ZNF638, but also to facilitate the transcription of ZNF638 via the stabilization of cAMP responsive element binding protein (CREB). USP7/ZNF638 axis selectively increased the cleavage of sterol regulatory element binding protein (SREBP1C) through AKT/mTORC1/S6K signaling, and formed USP7/ZNF638/SREBP1C nuclear complex to regulate lipogenesis-associated enzymes, including acetyl-CoA carboxylase (ACACA), fatty acid synthase (FASN), and Stearoyl-CoA desaturase (SCD). In the mice liver steatosis model induced by fructose, USP7 or ZNF638 abrogation significantly ameliorated disease progression. Furthermore, USP7/ZNF638 axis participated in the progression of lipogenesis-associated HCC. Our results have uncovered a novel mechanism of hepatic DNL, which might be beneficial to the development of new therapeutic targets for hepatic lipogenesis-associated diseases.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures





References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

