Structural Basis of SARS-CoV-2 and SARS-CoV Antibody Interactions
- PMID: 33041212
- PMCID: PMC7498231
- DOI: 10.1016/j.it.2020.09.004
Structural Basis of SARS-CoV-2 and SARS-CoV Antibody Interactions
Abstract
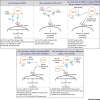
The 2019 coronavirus pandemic remains a major public health concern. Neutralizing antibodies (nAbs) represent a cutting-edge antiviral strategy. We focus here on severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and SARS-CoV, and discuss current progress in antibody research against rampant SARS-CoV-2 infections. We provide a perspective on the mechanisms of SARS-CoV-2-derived nAbs, comparing these with existing SARS-CoV-derived antibodies. We offer insight into how these antibodies cross-react and cross-neutralize by analyzing available structures of spike (S) glycoprotein-antibody complexes. We also propose ways of adopting antibody-based strategies - such as cocktail antibody therapeutics against SARS-CoV-2 - to overcome the possible resistance of currently identified mutants and mitigate possible antibody-dependent enhancement (ADE) pathologies. This review provides a platform for the progression of antibody and vaccine design against SARS-CoV-2, and possibly against future coronavirus pandemics.
Keywords: COVID-19; S glycoprotein; SARS-CoV; SARS-CoV-2; antibodies; hACE-2.
Copyright © 2020 Elsevier Ltd. All rights reserved.
Figures


References
-
- Liu Y., et al. Aerodynamic analysis of SARS-CoV-2 in two Wuhan hospitals. Nature. 2020;582:557–560. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

