NWB Query Engines: Tools to Search Data Stored in Neurodata Without Borders Format
- PMID: 33041776
- PMCID: PMC7526650
- DOI: 10.3389/fninf.2020.00027
NWB Query Engines: Tools to Search Data Stored in Neurodata Without Borders Format
Abstract
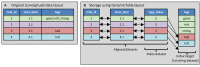
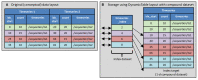
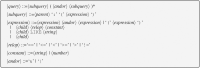
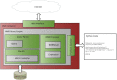
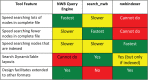
The Neurodata Without Borders (abbreviation NWB) format is a current technology for storing neurophysiology data along with the associated metadata. Data stored in the format is organized into separate HDF5 files, each file usually storing the data associated with a single recording session. While the NWB format provides a structured method for storing data, so far there have not been tools which enable searching a collection of NWB files in order to find data of interest for a particular purpose. We describe here three tools to enable searching NWB files. The tools have different features making each of them most useful for a particular task. The first tool, called the NWB Query Engine, is written in Java. It allows searching the complete content of NWB files. It was designed for the first version of NWB (NWB 1) and supports most (but not all) features of the most recent version (NWB 2). For some searches, it is the fastest tool. The second tool, called "search_nwb" is written in Python and also allow searching the complete contents of NWB files. It works with both NWB 1 and NWB 2, as does the third tool. The third tool, called "nwbindexer" enables searching a collection of NWB files using a two-step process. In the first step, a utility is run which creates an SQLite database containing the metadata in a collection of NWB files. This database is then searched in the second step, using another utility. Once the index is built, this two-step processes allows faster searches than are done by the other tools, but does not enable as complete of searches. All three tools use a simple query language which was developed for this project. Software integrating the three tools into a web-interface is provided which enables searching NWB files by submitting a web form.
Keywords: HDF5; Java; NWB format; Python; SQLite; metadata; neurophysiology; search.
Copyright © 2020 Ježek, Teeters and Sommer.
Figures





Similar articles
-
The Neurodata Without Borders ecosystem for neurophysiological data science.Elife. 2022 Oct 4;11:e78362. doi: 10.7554/eLife.78362. Elife. 2022. PMID: 36193886 Free PMC article.
-
Core principles for the implementation of the neurodata without borders data standard.J Neurosci Methods. 2021 Jan 15;348:108972. doi: 10.1016/j.jneumeth.2020.108972. Epub 2020 Nov 4. J Neurosci Methods. 2021. PMID: 33157146
-
A NWB-based dataset and processing pipeline of human single-neuron activity during a declarative memory task.Sci Data. 2020 Mar 4;7(1):78. doi: 10.1038/s41597-020-0415-9. Sci Data. 2020. PMID: 32132545 Free PMC article.
-
Image file formats: past, present, and future.Radiographics. 2001 May-Jun;21(3):789-98. doi: 10.1148/radiographics.21.3.g01ma25789. Radiographics. 2001. PMID: 11353125 Review.
-
A Systematic Review of Negative Work Behavior: Toward an Integrated Definition.Front Psychol. 2021 Oct 27;12:726973. doi: 10.3389/fpsyg.2021.726973. eCollection 2021. Front Psychol. 2021. PMID: 34777108 Free PMC article.
Cited by
-
Project, toolkit, and database of neuroinformatics ecosystem: A summary of previous studies on "Frontiers in Neuroinformatics".Front Neuroinform. 2022 Sep 26;16:902452. doi: 10.3389/fninf.2022.902452. eCollection 2022. Front Neuroinform. 2022. PMID: 36225654 Free PMC article. Review.
References
-
- Chou J., Howison M., Austin B., Wu K., Qiang J., Bethel E. W., et al. (2011). Parallel index and query for large scale data analysis, in 2011 International Conference for High Performance Computing, Networking, Storage and Analysis (SC) (Seattle, WA: ), 1–11. 10.1145/2063384.2063424 - DOI
-
- Folk M., Heber G., Koziol Q., Pourmal E., Robinson D. (2011). An overview of the HDF5 technology suite and its applications, in Proceedings of the EDBT/ICDT 2011 Workshop on Array Databases, AD '11 (New York, NY: ACM; ), 36–47. 10.1145/1966895.1966900 - DOI
LinkOut - more resources
Full Text Sources

