Integrating Machine Learning-Based Virtual Screening With Multiple Protein Structures and Bio-Assay Evaluation for Discovery of Novel GSK3β Inhibitors
- PMID: 33041806
- PMCID: PMC7517831
- DOI: 10.3389/fphar.2020.566058
Integrating Machine Learning-Based Virtual Screening With Multiple Protein Structures and Bio-Assay Evaluation for Discovery of Novel GSK3β Inhibitors
Abstract
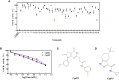
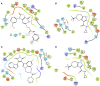
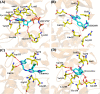
Glycogen synthase kinase-3β (GSK3β) is associated with various key biological processes, and it has been considered as a critical therapeutic target for the treatment of many diseases. However, it is a big challenge to develop ATP-competition GSK3β inhibitors because of the high sequence homology with other kinases. In this work, a novel parallel virtual screening strategy based on multiple GSK3β protein structures, integrating molecular docking, complex-based pharmacophore, and naive Bayesian classification, was developed to screen a large chemical database, the 50 compounds with top-scores then underwent a luminescent kinase assay, which led to the discovery of two GSK3β inhibitor hits. The high screening enrichment rate indicates the reliability and practicability of the integrated protocol. Finally, molecular docking and molecular dynamics simulation were employed to investigate the binding modes of the GSK3β inhibitors, and some "hot residues" critical to GSK3β affinity were highlighted. The present study may provide some valuable guidance for the development of novel GSK3β inhibitors.
Keywords: GSK3β; glycogen synthase kinase-3 beta inhibitor; molecular docking; molecular dynamics simulation; naive Bayesian classification; pharmacophore; virtual screening.
Copyright © 2020 Zhu, Wu, Wang, Li, Xu, Chen, Cai and Jin.
Figures



Similar articles
-
Theoretical Studies on the Selectivity Mechanisms of Glycogen Synthase Kinase 3β (GSK3β) with Pyrazine ATP-competitive Inhibitors by 3DQSAR, Molecular Docking, Molecular Dynamics Simulation and Free Energy Calculations.Curr Comput Aided Drug Des. 2020;16(1):17-30. doi: 10.2174/1573409915666190708102459. Curr Comput Aided Drug Des. 2020. PMID: 31284868 Free PMC article.
-
Discovery of novel selective PI3Kγ inhibitors through combining machine learning-based virtual screening with multiple protein structures and bio-evaluation.J Adv Res. 2021 Apr 20;36:1-13. doi: 10.1016/j.jare.2021.04.007. eCollection 2022 Feb. J Adv Res. 2021. PMID: 35127160 Free PMC article.
-
Identification of New GSK3β Inhibitors through a Consensus Machine Learning-Based Virtual Screening.Int J Mol Sci. 2023 Dec 7;24(24):17233. doi: 10.3390/ijms242417233. Int J Mol Sci. 2023. PMID: 38139062 Free PMC article.
-
Optimization of virtual screening against phosphoinositide 3-kinase delta: Integration of common feature pharmacophore and multicomplex-based molecular docking.Comput Biol Chem. 2024 Apr;109:108011. doi: 10.1016/j.compbiolchem.2023.108011. Epub 2024 Jan 2. Comput Biol Chem. 2024. PMID: 38198965
-
An emerging strategy for cancer treatment targeting aberrant glycogen synthase kinase 3 beta.Anticancer Agents Med Chem. 2009 Dec;9(10):1114-22. doi: 10.2174/187152009789734982. Anticancer Agents Med Chem. 2009. PMID: 19925395 Review.
Cited by
-
New Advances in the Pharmacology and Toxicology of Lithium: A Neurobiologically Oriented Overview.Pharmacol Rev. 2024 May 2;76(3):323-357. doi: 10.1124/pharmrev.120.000007. Pharmacol Rev. 2024. PMID: 38697859 Free PMC article. Review.
-
Discovery of Novel c-Jun N-Terminal Kinase 1 Inhibitors from Natural Products: Integrating Artificial Intelligence with Structure-Based Virtual Screening and Biological Evaluation.Molecules. 2022 Sep 22;27(19):6249. doi: 10.3390/molecules27196249. Molecules. 2022. PMID: 36234788 Free PMC article.
-
TargIDe: a machine-learning workflow for target identification of molecules with antibiofilm activity against Pseudomonas aeruginosa.J Comput Aided Mol Des. 2023 Jun;37(5-6):265-278. doi: 10.1007/s10822-023-00505-5. Epub 2023 Apr 22. J Comput Aided Mol Des. 2023. PMID: 37085636 Free PMC article.
-
In Silico Prediction of New Inhibitors for Kirsten Rat Sarcoma G12D Cancer Drug Target Using Machine Learning-Based Virtual Screening, Molecular Docking, and Molecular Dynamic Simulation Approaches.Pharmaceuticals (Basel). 2024 Apr 25;17(5):551. doi: 10.3390/ph17050551. Pharmaceuticals (Basel). 2024. PMID: 38794122 Free PMC article.
-
Identification of novel covalent JAK3 inhibitors through consensus scoring virtual screening: integration of common feature pharmacophore and covalent docking.Mol Divers. 2025 Apr;29(2):1353-1373. doi: 10.1007/s11030-024-10918-5. Epub 2024 Jul 15. Mol Divers. 2025. PMID: 39009908
References
-
- Atkinson J. M., Rank K. B., Zeng Y., Capen A., Yadav V., Manro J. R., et al. (2015). Activating the Wnt/beta-Catenin Pathway for the Treatment of Melanoma–Application of LY2090314, a Novel Selective Inhibitor of Glycogen Synthase Kinase-3. PloS One 10 (4), e0125028. 10.1371/journal.pone.0125028 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources

