High-Throughput Sequencing for Deciphering the Virome of Alfalfa (Medicago sativa L.)
- PMID: 33042059
- PMCID: PMC7518122
- DOI: 10.3389/fmicb.2020.553109
High-Throughput Sequencing for Deciphering the Virome of Alfalfa (Medicago sativa L.)
Abstract
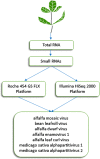
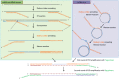
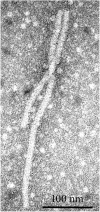
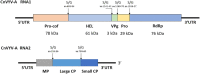
Alfalfa (Medicago sativa L.), also known as lucerne, is a major forage crop worldwide. In the United States, it has recently become the third most valuable field crop, with an estimated value of over $9.3 billion. Alfalfa is naturally infected by many different pathogens, including viruses, obligate parasites that reproduce only inside living host cells. Traditionally, viral infections of alfalfa have been considered by breeders, growers, producers and researchers to be diseases of limited importance, although they are widespread in all major cultivation areas. However, over the past few years, due to the rapid development of high-throughput sequencing (HTS), viral metagenomics, bioinformatics tools for interpreting massive amounts of HTS data and the increasing accessibility of public data repositories for transcriptomic discoveries, several emerging viruses of alfalfa with the potential to cause serious yield losses have been described. They include alfalfa leaf curl virus (family Geminiviridae), alfalfa dwarf virus (family Rhabdoviridae), alfalfa enamovirus 1 (family Luteoviridae), alfalfa virus S (family Alphaflexiviridae) and others. These discoveries have called into question the assumed low economic impact of viral diseases in alfalfa and further suggested their possible contribution to the severity of complex infections involving multiple pathogens. In this review, we will focus on viruses of alfalfa recently described in different laboratories on the basis of the above research methodologies.
Keywords: Medicago sativa L.; alfalfa; emerging viruses; high throughput sequencing; virome.
Copyright © 2020 Bejerman, Roumagnac and Nemchinov.
Figures






References
-
- Alliot B., Signoret P. A. (1972). La maladie a enations de la lucerne, une maladie nouvelle pour la France. Phytopathol. Z 74:69 10.1111/j.1439-0434.1972.tb04647.x - DOI
-
- Babadoost M. (1990). Virus Diseases of Alfalfa and Clovers in Illinois. Reports on Plant Diseases, RPD No. 307, Champaign: University of Illinois at Urbana-Champaign.
Publication types
LinkOut - more resources
Full Text Sources
Miscellaneous

