Salinity Stress Enhances the Antioxidant Capacity of Bacillus and Planococcus Species Isolated From Saline Lake Environment
- PMID: 33042068
- PMCID: PMC7521018
- DOI: 10.3389/fmicb.2020.561816
Salinity Stress Enhances the Antioxidant Capacity of Bacillus and Planococcus Species Isolated From Saline Lake Environment
Abstract
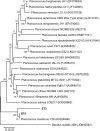
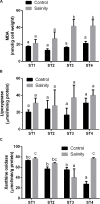
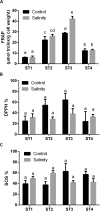
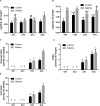
This study aims at exploiting salinity stress as an innovative, simple, and cheap method to enhance the production of antioxidant metabolites and enzymes from bacteria for potential application as functional additives to foods and pharmaceuticals. We investigated the physiological and biochemical responses of four bacterial isolates, which exhibited high tolerance to 20% NaCl (wt/vol), out of 27 bacterial strains isolated from Aushazia Lake, Qassim region, Saudi Arabia. The phylogenetic analysis of the 16S rRNA genes of these four isolates indicated that strains ST1 and ST2 belong to genus Bacillus, whereas strains ST3 and ST4 belong to genus Planococcus. Salinity stress differentially induced oxidative damage, where strains ST3 and ST4 showed increased lipid peroxidation, lipoxygenase, and xanthine oxidase levels. Consequently, high antioxidant contents were produced to control oxidative stress, particularly in ST3 and ST4. These two Planococcus strains showed increased glutathione cycle, phenols, flavonoids, antioxidant capacity, catalase, and/or superoxide dismutase (SOD). Interestingly, the production of glutathione by Planococcus strains was some thousand folds greater than by higher plants. On the other hand, the induction of antioxidants in ST1 and ST2 was restricted to phenols, flavonoids, peroxidase, glutaredoxin, and/or SOD. The hierarchical analysis also supported strain-specific responses. This is the first report that exploited salinity stress for promoting the production of antioxidants from bacterial isolates, which can be utilized as postbiotics for promising applications in foods and pharmaceuticals.
Keywords: Bacillus; Planococcus; antioxidants; salinity stress; stress markers.
Copyright © 2020 Hassan, Alkhalifah, Al Yousef, Beemster, Mousa, Hozzein and AbdElgawad.
Figures



Similar articles
-
Heat stress as an innovative approach to enhance the antioxidant production in Pseudooceanicola and Bacillus isolates.Sci Rep. 2020 Sep 15;10(1):15076. doi: 10.1038/s41598-020-72054-y. Sci Rep. 2020. PMID: 32934293 Free PMC article.
-
Bacillus firmus (SW5) augments salt tolerance in soybean (Glycine max L.) by modulating root system architecture, antioxidant defense systems and stress-responsive genes expression.Plant Physiol Biochem. 2018 Nov;132:375-384. doi: 10.1016/j.plaphy.2018.09.026. Epub 2018 Sep 21. Plant Physiol Biochem. 2018. PMID: 30268029
-
Nutritional Status as the Key Modulator of Antioxidant Responses Induced by High Environmental Ammonia and Salinity Stress in European Sea Bass (Dicentrarchus labrax).PLoS One. 2015 Aug 4;10(8):e0135091. doi: 10.1371/journal.pone.0135091. eCollection 2015. PLoS One. 2015. PMID: 26241315 Free PMC article.
-
Interactive effects of silicon and arbuscular mycorrhiza in modulating ascorbate-glutathione cycle and antioxidant scavenging capacity in differentially salt-tolerant Cicer arietinum L. genotypes subjected to long-term salinity.Protoplasma. 2016 Sep;253(5):1325-45. doi: 10.1007/s00709-015-0892-4. Epub 2015 Oct 14. Protoplasma. 2016. PMID: 26468060
-
The role of antioxidant responses on the tolerance range of extreme halophyte Salsola crassa grown under toxic salt concentrations.Ecotoxicol Environ Saf. 2014 Dec;110:21-30. doi: 10.1016/j.ecoenv.2014.08.013. Epub 2014 Sep 3. Ecotoxicol Environ Saf. 2014. PMID: 25193881
Cited by
-
An Isolated Arthrobacter sp. Enhances Rice (Oryza sativa L.) Plant Growth.Microorganisms. 2022 Jun 9;10(6):1187. doi: 10.3390/microorganisms10061187. Microorganisms. 2022. PMID: 35744704 Free PMC article.
-
Haloadaptative Responses of Aspergillus sydowii to Extreme Water Deprivation: Morphology, Compatible Solutes, and Oxidative Stress at NaCl Saturation.J Fungi (Basel). 2020 Nov 27;6(4):316. doi: 10.3390/jof6040316. J Fungi (Basel). 2020. PMID: 33260894 Free PMC article.
-
Genomic and Physiological Characterization of Bacilli Isolated From Salt-Pans With Plant Growth Promoting Features.Front Microbiol. 2021 Sep 13;12:715678. doi: 10.3389/fmicb.2021.715678. eCollection 2021. Front Microbiol. 2021. PMID: 34589073 Free PMC article.
-
Novel Halotolerant Bacteria from Saline Environments: Isolation and Biomolecule Production.BioTech (Basel). 2025 Jun 19;14(2):49. doi: 10.3390/biotech14020049. BioTech (Basel). 2025. PMID: 40558398 Free PMC article.
-
The identification of the new species Nitratireductor thuwali sp. nov. reveals the untapped diversity of hydrocarbon-degrading culturable bacteria from the arid mangrove sediments of the Red Sea.Front Microbiol. 2023 May 2;14:1155381. doi: 10.3389/fmicb.2023.1155381. eCollection 2023. Front Microbiol. 2023. PMID: 37200916 Free PMC article.
References
-
- Abdelaal K. A., El-Maghraby L. M., Elansary H., Hafez Y. M., Ibrahim E. I., El-Banna M., et al. (2020). Treatment of sweet pepper with stress tolerance-inducing compounds alleviates salinity stress oxidative damage by mediating the physio-biochemical activities and antioxidant systems. Agronomy 10:26 10.3390/agronomy10010026 - DOI
-
- AbdElgawad H., De Vos D., Zinta G., Domagalska M. A., Beemster G. T. S., Asard H. (2015). Grassland species differentially regulate proline concentrations under future climate conditions: an integrated biochemical and modelling approach. New Phytol. 208 354–369. 10.1111/nph.13481 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

