Analysis of mesenchymal stem cell proteomes in situ in the ischemic heart
- PMID: 33042285
- PMCID: PMC7532665
- DOI: 10.7150/thno.47893
Analysis of mesenchymal stem cell proteomes in situ in the ischemic heart
Abstract
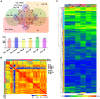
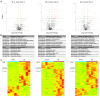
Rationale: Cell therapy for myocardial infarction is promising but largely unsuccessful in part due to a lack of mechanistic understanding. Techniques enabling identification of stem cell-specific proteomes in situ in the injured heart may shed light on how the administered cells respond to the injured microenvironment and exert reparative effects. Objective: To identify the proteomes of the transplanted mesenchymal stem cells (MSCs) in the infarcted myocardium, we sought to target a mutant methionyl-tRNA synthetase (MetRSL274G) in MSCs, which charges azidonorleucine (ANL), a methionine analogue and non-canonical amino acid, to tRNA and subsequently to nascent proteins, permitting isolation of ANL-labeled MSC proteomes from ischemic hearts by ANL-alkyne based click reaction. Methods and Results: Murine MSCs were transduced with lentivirus MetRSL274G and supplemented with ANL; the ANL-tagged nascent proteins were visualized by bio-orthogonal non-canonical amino-acid tagging, spanning all molecular weights and by fluorescent non-canonical amino-acid tagging, displaying strong fluorescent signal. Then, the MetRSL274G-transduced MSCs were administered to the infarcted or Sham heart in mice receiving ANL treatment. The MSC proteomes were isolated from the left ventricular protein lysates by click reaction at days 1, 3, and 7 after cell administration, identified by LC/MS. Among all identified proteins (in Sham and MI hearts, three time-points each), 648 were shared by all 6 groups, accounting for 82±5% of total proteins in each group, and enriched under mitochondrion, extracellular exosomes, oxidation-reduction process and poly(A) RNA binding. Notably, 26, 110 and 65 proteins were significantly up-regulated and 11, 28 and 19 proteins were down-regulated in the infarcted vs. Sham heart at the three time-points, respectively; these proteins are pronounced in the GO terms of extracellular matrix organization, response to stress and regulation of apoptotic process and in the KEGG pathways of complements and coagulation cascades, apoptosis, and regulators of actin cytoskeleton. Conclusions: MetRSL274G expression allows successful identification of MSC-specific nascent proteins in the infarcted hearts, which reflect the functional states, adaptive response, and reparative effects of MSCs that may be leveraged to improve cardiac repair.
Keywords: Methionyl-tRNA synthetase; mass spectrometry; mesenchymal stem cells; myocardial infarction; proteomics.
© The author(s).
Conflict of interest statement
Competing Interests: The authors have declared that no competing interest exists.
Figures





References
-
- Tao B, Cui M, Wang C, Ma S, Wu F, Yi F. et al. Percutaneous intramyocardial delivery of mesenchymal stem cells induces superior improvement in regional left ventricular function compared with bone marrow mononuclear cells in porcine myocardial infarcted heart. Theranostics. 2015;5:196–205. - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials

