Intramuscular accumulation of pentadecanoic acid activates AKT1 to phosphorylate NCOR1 and triggers FOXM1-mediated apoptosis in the pathogenesis of sarcopenia
- PMID: 33042406
- PMCID: PMC7540105
Intramuscular accumulation of pentadecanoic acid activates AKT1 to phosphorylate NCOR1 and triggers FOXM1-mediated apoptosis in the pathogenesis of sarcopenia
Abstract
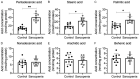
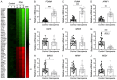
Sarcopenia is an age-associated disorder that results in skeletal muscle loss. Apoptosis and inflammation are the two major contributors to sarcopenia. Emerging evidence has shown that long-chain fatty acids (LCFAs) are implicated in the muscles of sarcopenic animal models. However, it is unknown whether LCFAs are correlated with apoptosis or inflammation in the pathogenesis of sarcopenia. Herein, we found that pentadecanoic acid (PDA), a C15 LCFA, was significantly accumulated in human sarcopenic muscles. In vitro PDA treatment could dose-dependently induce the expression of the transcription factor FOXM1 (forkhead box M1) and several proapoptotic genes, such as PUMA (p53-upregulated modulator of apoptosis), BAX (B-cell/lymphoma 2-associated X) and APAF1 (apoptotic peptidase activating factor 1), thereby causing apoptosis. Mechanically, PDA activated AKT1 (AKT serine/threonine kinase 1) to phosphorylate NCOR1 (nuclear receptor corepressor 1). The phosphorylated NCOR1 disassociated from the NCOR1-FOXM1 transcriptional complex and could not repress FOXM1-mediated transcription, leading to the induction of PUMA. The activated PUMA further triggered downstream apoptotic signaling, including activation of the BAX, APAF1 and caspase cascades, leading to the occurrence of apoptosis. Alkaline phosphatase or knockdown of AKT1 in vitro reversed the FOXM1-mediated apoptotic signaling. Collectively, our results provide new evidence that LCFAs are involved in the pathogenesis of sarcopenia by activating apoptotic signaling. Attempts to decrease the intake of PDA-containing foods or blocking AKT1 may improve the symptoms of sarcopenia.
Keywords: AKT1; FOXM1; NCOR1; Sarcopenia; apoptosis; pentadecanoic acid.
AJTR Copyright © 2020.
Conflict of interest statement
None.
Figures







References
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous
