Choice of Nanovaccine Delivery Mode Has Profound Impacts on the Intralymph Node Spatiotemporal Distribution and Immunotherapy Efficacy
- PMID: 33042743
- PMCID: PMC7539204
- DOI: 10.1002/advs.202001108
Choice of Nanovaccine Delivery Mode Has Profound Impacts on the Intralymph Node Spatiotemporal Distribution and Immunotherapy Efficacy
Abstract
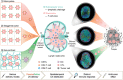
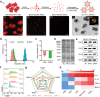
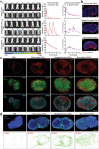
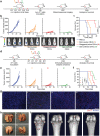
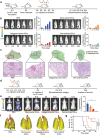
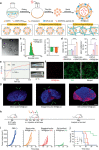
Nanovaccines have attracted booming interests in vaccinology studies, but the profound impacts of their delivery mode on immune response remain unrealized. Herein, immunostimulatory CpG-modified tumor-derived nanovesicles (CNVs) are used as a nanovaccine testbed to initially evaluate the impacts of three distinct delivery modes, including mono-pulse CNVs, staggered-pulse CNVs, and gel-confined CNVs. Fundamentally, delivery mode has enormous impacts on the immunomodulatory effects, altering the spatiotemporal distribution of nanovaccine residence and dendritic cell-T cell interaction in lymph nodes, and finally affecting subsequent T cell-mediated immune performance. As a result, the gel-confined delivery mode offers the best therapeutic performance in multiple tumor models. When extending evaluation to examine how the various delivery modes impact the performance of liposome-based nanovaccines, similar trends in intralymph node distribution and antitumor effect are observed. This work provides a strong empirical foundation that nanovaccine researchers should position delivery mode near the top of their considerations for the experimental design, which should speed up nanovaccine development and facilitate efficient selection of appropriate delivery modes in the clinic.
Keywords: delivery modes; hydrogels; lymph nodes; nanovaccines; tumor immunotherapy.
© 2020 The Authors. Published by Wiley‐VCH GmbH.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Similar articles
-
CpG-Based Nanovaccines for Cancer Immunotherapy.Int J Nanomedicine. 2021 Aug 5;16:5281-5299. doi: 10.2147/IJN.S317626. eCollection 2021. Int J Nanomedicine. 2021. PMID: 34385817 Free PMC article. Review.
-
Nanovaccine-based strategies for lymph node targeted delivery and imaging in tumor immunotherapy.J Nanobiotechnology. 2023 Jul 23;21(1):236. doi: 10.1186/s12951-023-01989-x. J Nanobiotechnology. 2023. PMID: 37482608 Free PMC article. Review.
-
Facile preparation of a metal-phenolic network-based lymph node targeting nanovaccine for antitumor immunotherapy.Acta Biomater. 2023 Mar 1;158:510-524. doi: 10.1016/j.actbio.2022.12.066. Epub 2023 Jan 2. Acta Biomater. 2023. PMID: 36603733
-
Intratumoral administration of STING-activating nanovaccine enhances T cell immunotherapy.J Immunother Cancer. 2022 May;10(5):e003960. doi: 10.1136/jitc-2021-003960. J Immunother Cancer. 2022. PMID: 35623658 Free PMC article.
-
Manganese oxide-constructed multifunctional biomimetic nanovaccine for robust tumor-specific T cell priming and chemodynamic therapy.Biomaterials. 2024 Sep;309:122626. doi: 10.1016/j.biomaterials.2024.122626. Epub 2024 May 20. Biomaterials. 2024. PMID: 38795524
Cited by
-
CpG-Based Nanovaccines for Cancer Immunotherapy.Int J Nanomedicine. 2021 Aug 5;16:5281-5299. doi: 10.2147/IJN.S317626. eCollection 2021. Int J Nanomedicine. 2021. PMID: 34385817 Free PMC article. Review.
-
Nanovaccine-based strategies for lymph node targeted delivery and imaging in tumor immunotherapy.J Nanobiotechnology. 2023 Jul 23;21(1):236. doi: 10.1186/s12951-023-01989-x. J Nanobiotechnology. 2023. PMID: 37482608 Free PMC article. Review.
-
Application of Nano-Delivery Systems in Lymph Nodes for Tumor Immunotherapy.Nanomicro Lett. 2023 Jun 3;15(1):145. doi: 10.1007/s40820-023-01125-2. Nanomicro Lett. 2023. PMID: 37269391 Free PMC article. Review.
-
CD47KO/CRT dual-bioengineered cell membrane-coated nanovaccine combined with anti-PD-L1 antibody for boosting tumor immunotherapy.Bioact Mater. 2022 Oct 4;22:211-224. doi: 10.1016/j.bioactmat.2022.09.017. eCollection 2023 Apr. Bioact Mater. 2022. PMID: 36246666 Free PMC article.
-
Three-dimensional (3D) scaffolds as powerful weapons for tumor immunotherapy.Bioact Mater. 2022 Jan 26;17:300-319. doi: 10.1016/j.bioactmat.2022.01.020. eCollection 2022 Nov. Bioact Mater. 2022. PMID: 35386452 Free PMC article. Review.
References
LinkOut - more resources
Full Text Sources
Research Materials
