Assessing the SARS-CoV-2 threat to wildlife: Potential risk to a broad range of mammals
- PMID: 33043253
- PMCID: PMC7534737
- DOI: 10.1016/j.pecon.2020.09.008
Assessing the SARS-CoV-2 threat to wildlife: Potential risk to a broad range of mammals
Abstract
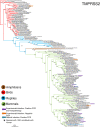
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) can infect animals, however, the whole range of potential hosts is still unknown. This work makes an assessment of wildlife susceptibility to SARS-CoV-2 by analyzing the similarities of Angiotensin Converting Enzyme 2 (ACE2) and Transmembrane Protease, Serine 2 (TMPRSS2)-both recognized as receptors and protease for coronavirus spike protein-and the genetic variation of the viral protein spike in the recognition sites. The sequences from different mammals, birds, reptiles, and amphibians, and the sequence from SARS-CoV-2 S protein were obtained from the GenBank. Comparisons of aligned sequences were made by selecting amino acids residues of ACE2, TMPRSS2 and S protein; phylogenetic trees were reconstructed using the same sequences. The species susceptibility was ranked by substituting the values of amino acid residues for both proteins. Our results ranked primates at the top, but surprisingly, just below are carnivores, cetaceans and wild rodents, showing a relatively high potential risk, as opposed to lab rodents that are typically mammals at lower risk. Most of the sequences from birds, reptiles and amphibians occupied the lowest ranges in the analyses. Models and phylogenetic trees outputs showed the species that are more prone to getting infected with SARS-CoV-2. Interestingly, during this short pandemic period, a high haplotypic variation was observed in the RBD of the viral S protein, suggesting new risks for other hosts. Our findings are consistent with other published results reporting laboratory and natural infections in different species. Finally, urgent measures of wildlife monitoring are needed regarding SARS-CoV-2, as well as measures for avoiding or limiting human contact with wildlife, and precautionary measures to protect wildlife workers and researchers; monitoring disposal of waste and sewage than can potentially affect the environment, and designing protocols for dealing with the outbreak.
Keywords: COVID-19; Carnivores; Cetacean; Rodent; SARS-CoV-2; Wildlife.
© 2020 Associação Brasileira de Ciência Ecológica e Conservação. Published by Elsevier B.V.
Figures




References
-
- APHIS-USDA . 2020. Confirmation of COVID-19 in Two Pet Cats in New York. https://www.aphis.usda.gov/aphis/newsroom/news/sa_by_date/sa-2020/sars-c... (accessed 09.05.20)
-
- Ahmed W., Angel N., Edson J., Bibby K., Bivins A., O’Brien J., Choi P., Kitajima M., Simpson S., Li J., Tscharke B., Verhagen R., Smith W., Zaugg J., Dierens L., Hugenholtz P., Thomas K., Mueller J. First confirmed detection of SARS-CoV-2 in untreated wastewater in Australia: a proof of concept for the wastewater surveillance of COVID-19 in the community. Sci. Total Environ. 2020;728:138764. doi: 10.1016/j.scitotenv.2020.138764. - DOI - PMC - PubMed
-
- Bao L., Deng W., Gao H., Xiao C., Liu J., Xue J., Lv Q., Liu J., Yu P., Xu Y., Qi F., Qu Y., Li F., Xiang Z., Yu H., Gong S., Liu M., Wang G., Wang S., Song Z., Zhao W., Han Y., Zhao L., Liu X., Wei Q., Qin C. Reinfection could not occur in SARS-CoV-2 infected rhesus macaques. BioRxiv. 2020 doi: 10.1101/2020.03.13.990226. - DOI
Publication types
LinkOut - more resources
Full Text Sources
Miscellaneous
