Furin: A Potential Therapeutic Target for COVID-19
- PMID: 33043282
- PMCID: PMC7534598
- DOI: 10.1016/j.isci.2020.101642
Furin: A Potential Therapeutic Target for COVID-19
Abstract
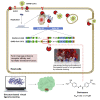
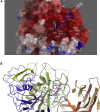
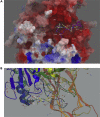
COVID-19 broke out in the end of December 2019 and is still spreading rapidly, which has been listed as an international concerning public health emergency. We found that the Spike protein of SARS-CoV-2 contains a furin cleavage site, which did not exist in any other betacoronavirus subtype B. Based on a series of analysis, we speculate that the presence of a redundant furin cut site in its Spike protein is responsible for SARS-CoV-2's stronger infectious nature than other coronaviruses, which leads to higher membrane fusion efficiency. Subsequently, a library of 4,000 compounds including approved drugs and natural products was screened against furin through structure-based virtual screening and then assayed for their inhibitory effects on furin activity. Among them, an anti-parasitic drug, diminazene, showed the highest inhibition effects on furin with an IC50 of 5.42 ± 0.11 μM, which might be used for the treatment of COVID-19.
Keywords: Computational Molecular Modelling; Disease; Drugs; Virology.
© 2020 The Author(s).
Conflict of interest statement
Authors declare no conflict of interest and no competing financial interest.
Figures


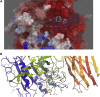
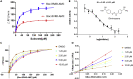
References
-
- Dahms S.O., Hardes K., Steinmetzer T., Than M.E. X-ray Structures of the proprotein convertase furin bound with substrate analogue inhibitors reveal substrate specificity determinants beyond the S4 pocket. Biochemistry. 2018;57:925–934. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

