Inhibition of Influenza Virus Polymerase by Interfering with Its Protein-Protein Interactions
- PMID: 33044059
- PMCID: PMC8204303
- DOI: 10.1021/acsinfecdis.0c00552
Inhibition of Influenza Virus Polymerase by Interfering with Its Protein-Protein Interactions
Abstract
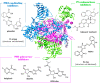
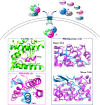
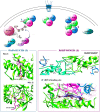
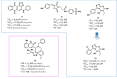
Influenza (flu) virus is a serious threat to global health with the potential to generate devastating pandemics. The availability of broad spectrum antiviral drugs is an unequaled weapon during pandemic events, especially when a vaccine is still not available. One of the most promising targets for the development of new antiflu therapeutics is the viral RNA-dependent RNA polymerase (RdRP). The assembly of the flu RdRP heterotrimeric complex from the individual polymerase acidic protein (PA), polymerase basic protein 1 (PB1), and polymerase basic protein 2 (PB2) subunits is a prerequisite for RdRP functions, such as mRNA synthesis and genome replication. In this Review, we report the known protein-protein interactions (PPIs) occurring by RdRP that could be disrupted by small molecules and analyze their benefits and drawbacks as drug targets. An overview of small molecules able to interfere with flu RdRP functions exploiting the PPI inhibition approach is described. In particular, an update on the most recent inhibitors targeting the well-consolidated RdRP PA-PB1 subunit heterodimerization is mainly reported, together with pioneer inhibitors targeting other virus-virus or virus-host interactions involving RdRP subunits. As demonstrated by the PA-PB1 interaction inhibitors discussed herein, the inhibition of flu RdRP functions by PPI disrupters clearly represents a valid means to identify compounds endowed with a broad spectrum of action and a reduced propensity to develop drug resistance, which are the main issues of antiviral drugs.
Keywords: PA−PB1; PA−Pol II CTD; PB1−PB2; PB1−RanBP5; PB2−ANP32; PB2−importin-α; RNA-dependent RNA polymerase; anti-influenza small molecules; protein−protein interface inhibitors.
Conflict of interest statement
The authors declare no competing financial interest.
Figures



References
-
- Garten R. J.; Davis C. T.; Russell C. A.; Shu B.; Lindstrom S.; Balish A.; Sessions W. M.; Xu X.; Skepner E.; Deyde V.; Okomo-Adhiambo M.; Gubareva L.; Barnes J.; Smith C. B.; Emery S. L.; Hillman M. J.; Rivailler P.; Smagala J.; De Graaf M.; Burke D. F.; Fouchier R. A. M.; Pappas C.; Alpuche-Aranda C. M.; López-Gatell H.; Olivera H.; López I.; Myers C. A.; Faix D.; Blair P. J.; Yu C.; Keene K. M.; Dotson P. D.; Boxrud D.; Sambol A. R.; Abid S. H.; St. George K.; Bannerman T.; Moore A. L.; Stringer D. J.; Blevins P.; Demmler-Harrison G. J.; Ginsberg M.; Kriner P.; Waterman S.; Smole S.; Guevara H. F.; Belongia E. A.; Clark P. A.; Beatrice S. T.; Donis R.; Katz J.; Finelli L.; Bridges C. B.; Shaw M.; Jernigan D. B.; Uyeki T. M.; Smith D. J.; Klimov A. I.; Cox N. J. (2009) Antigenic and Genetic Characteristics of Swine-Origin 2009 A(H1N1) Influenza Viruses Circulating in Humans. Science 325, 197–201. 10.1126/science.1176225. - DOI - PMC - PubMed
-
- World Health Organization (accessed 2020-09-04) Influenza, http://www.who.int/influenza.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

