CRISPR-based assays for rapid detection of SARS-CoV-2
- PMID: 33045362
- PMCID: PMC7546951
- DOI: 10.1016/j.ymeth.2020.10.003
CRISPR-based assays for rapid detection of SARS-CoV-2
Abstract
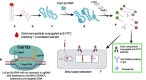
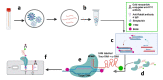
COVID-19 pandemic posed an unprecedented threat to global public health and economies. There is no effective treatment of the disease, hence, scaling up testing for rapid diagnosis of SARS-CoV-2 infected patients and quarantine them from healthy individuals is one the best strategies to curb the pandemic. Establishing globally accepted easy-to-access diagnostic tests is extremely important to understanding the epidemiology of the present pandemic. While nucleic acid based tests are considered to be more sensitive with respect to serological tests but present gold standard qRT-PCR-based assays possess limitations such as low sample throughput, requirement for sophisticated reagents and instrumentation. To overcome these shortcomings, recent efforts of incorporating LAMP-based isothermal detection, and minimizing the number of reagents required are on rise. CRISPR based novel techniques, when merge with isothermal and allied technologies, promises to provide sensitive and rapid detection of SARS-CoV-2 nucleic acids. Here, we discuss and present compilation of state-of-the-art detection techniques for COVID-19 using CRISPR technology which has tremendous potential to transform diagnostics and epidemiology.
Keywords: COVID-19; CRISPR diagnostics; Diagnostic advancements; Molecular testing; SARS-CoV-2.
Copyright © 2020 Elsevier Inc. All rights reserved.
Conflict of interest statement
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures




Similar articles
-
CLEVER assay: A visual and rapid RNA extraction-free detection of SARS-CoV-2 based on CRISPR-Cas integrated RT-LAMP technology.J Appl Microbiol. 2022 Aug;133(2):410-421. doi: 10.1111/jam.15571. Epub 2022 Apr 18. J Appl Microbiol. 2022. PMID: 35396760 Free PMC article.
-
High-Surety Isothermal Amplification and Detection of SARS-CoV-2.mSphere. 2021 May 19;6(3):e00911-20. doi: 10.1128/mSphere.00911-20. mSphere. 2021. PMID: 34011690 Free PMC article.
-
iSCAN: An RT-LAMP-coupled CRISPR-Cas12 module for rapid, sensitive detection of SARS-CoV-2.Virus Res. 2020 Oct 15;288:198129. doi: 10.1016/j.virusres.2020.198129. Epub 2020 Aug 18. Virus Res. 2020. PMID: 32822689 Free PMC article.
-
Strategies That Facilitate Extraction-Free SARS-CoV-2 Nucleic Acid Amplification Tests.Viruses. 2022 Jun 15;14(6):1311. doi: 10.3390/v14061311. Viruses. 2022. PMID: 35746782 Free PMC article. Review.
-
CRISPR-based tools: Alternative methods for the diagnosis of COVID-19.Clin Biochem. 2021 Mar;89:1-13. doi: 10.1016/j.clinbiochem.2020.12.011. Epub 2021 Jan 9. Clin Biochem. 2021. PMID: 33428900 Free PMC article. Review.
Cited by
-
Design and simulation of a millifluidic device for differential detection of SARS-CoV-2 and H1N1 based on triboelectricity.Bioelectrochemistry. 2022 Jun;145:108096. doi: 10.1016/j.bioelechem.2022.108096. Epub 2022 Mar 16. Bioelectrochemistry. 2022. PMID: 35316730 Free PMC article.
-
Application of CRISPR/Cas Systems in the Nucleic Acid Detection of Infectious Diseases.Diagnostics (Basel). 2022 Oct 11;12(10):2455. doi: 10.3390/diagnostics12102455. Diagnostics (Basel). 2022. PMID: 36292145 Free PMC article. Review.
-
Biochemical composition, transmission and diagnosis of SARS-CoV-2.Biosci Rep. 2021 Aug 27;41(8):BSR20211238. doi: 10.1042/BSR20211238. Biosci Rep. 2021. PMID: 34291285 Free PMC article. Review.
-
Creating respiratory pathogen-free environments in healthcare and nursing-care settings: a comprehensive review.Geroscience. 2025 Feb;47(1):543-571. doi: 10.1007/s11357-024-01379-7. Epub 2024 Oct 11. Geroscience. 2025. PMID: 39392557 Free PMC article. Review.
-
Current and innovative methods for the diagnosis of COVID‑19 infection (Review).Int J Mol Med. 2021 Jun;47(6):100. doi: 10.3892/ijmm.2021.4933. Epub 2021 Apr 13. Int J Mol Med. 2021. PMID: 33846767 Free PMC article. Review.
References
-
- Who.int, Coronavirus. (2020). https://www.who.int/emergencies/diseases/novel-coronavirus-2019 (accessed May 29, 2020).
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

