ViruSurf: an integrated database to investigate viral sequences
- PMID: 33045721
- PMCID: PMC7778888
- DOI: 10.1093/nar/gkaa846
ViruSurf: an integrated database to investigate viral sequences
Abstract
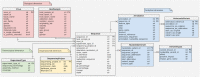
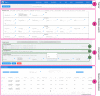
ViruSurf, available at http://gmql.eu/virusurf/, is a large public database of viral sequences and integrated and curated metadata from heterogeneous sources (RefSeq, GenBank, COG-UK and NMDC); it also exposes computed nucleotide and amino acid variants, called from original sequences. A GISAID-specific ViruSurf database, available at http://gmql.eu/virusurf_gisaid/, offers a subset of these functionalities. Given the current pandemic outbreak, SARS-CoV-2 data are collected from the four sources; but ViruSurf contains other virus species harmful to humans, including SARS-CoV, MERS-CoV, Ebola and Dengue. The database is centered on sequences, described from their biological, technological and organizational dimensions. In addition, the analytical dimension characterizes the sequence in terms of its annotations and variants. The web interface enables expressing complex search queries in a simple way; arbitrary search queries can freely combine conditions on attributes from the four dimensions, extracting the resulting sequences. Several example queries on the database confirm and possibly improve results from recent research papers; results can be recomputed over time and upon selected populations. Effective search over large and curated sequence data may enable faster responses to future threats that could arise from new viruses.
© The Author(s) 2020. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures

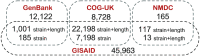
References
-
- Bernasconi A., Canakoglu A., Pinoli P., Ceri S.. Empowering Virus Sequences Research through Conceptual Modeling. 39th InternationalConference on Conceptual Model, Nov. 2020. 2020;
-
- Bernasconi A., Ceri S., Campi A., Masseroli M.. Mayr H.C., Guizzardi G., Ma H., Pastor O.. Conceptual Modeling for Genomics: Building an Integrated Repository of Open Data. Conceptual Modeling. 2017; Cham: Springer International Publishing; 325–339.
-
- Bernasconi A., Canakoglu A., Masseroli M., Ceri S.. META-BASE: a Novel Architecture for Large-Scale Genomic Metadata Integration. IEEE/ACM Trans. Comput. Biol. Bioinform. 2020; https://ieeexplore.ieee.org/document/9104916. - PubMed
-
- O’Leary N.A., Wright M.W., Brister J.R., Ciufo S., Haddad D., McVeigh R., Rajput B., Robbertse B., Smith-White B., Ako-Adjei D. et al. .. Reference sequence (RefSeq) database at NCBI: current status, taxonomic expansion, and functional annotation. Nucleic Acids Res. 2016; 44:D733–D745. - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

