GESS: a database of global evaluation of SARS-CoV-2/hCoV-19 sequences
- PMID: 33045727
- PMCID: PMC7778918
- DOI: 10.1093/nar/gkaa808
GESS: a database of global evaluation of SARS-CoV-2/hCoV-19 sequences
Abstract
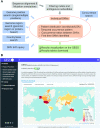
The COVID-19 outbreak has become a global emergency since December 2019. Analysis of SARS-CoV-2 sequences can uncover single nucleotide variants (SNVs) and corresponding evolution patterns. The Global Evaluation of SARS-CoV-2/hCoV-19 Sequences (GESS, https://wan-bioinfo.shinyapps.io/GESS/) is a resource to provide comprehensive analysis results based on tens of thousands of high-coverage and high-quality SARS-CoV-2 complete genomes. The database allows user to browse, search and download SNVs at any individual or multiple SARS-CoV-2 genomic positions, or within a chosen genomic region or protein, or in certain country/area of interest. GESS reveals geographical distributions of SNVs around the world and across the states of USA, while exhibiting time-dependent patterns for SNV occurrences which reflect development of SARS-CoV-2 genomes. For each month, the top 100 SNVs that were firstly identified world-widely can be retrieved. GESS also explores SNVs occurring simultaneously with specific SNVs of user's interests. Furthermore, the database can be of great help to calibrate mutation rates and identify conserved genome regions. Taken together, GESS is a powerful resource and tool to monitor SARS-CoV-2 migration and evolution according to featured genomic variations. It provides potential directive information for prevalence prediction, related public health policy making, and vaccine designs.
© The Author(s) 2020. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures



References
-
- Hu Y., Riley L.W.. Dissemination and co-circulation of SARS-CoV2 subclades exhibiting enhanced transmission associated with increased mortality in Western Europe and the United States. 2020; medRxiv doi:15 July 2020, preprint: not peer reviewed 10.1101/2020.07.13.20152959. - DOI
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

