Open Targets Genetics: systematic identification of trait-associated genes using large-scale genetics and functional genomics
- PMID: 33045747
- PMCID: PMC7778936
- DOI: 10.1093/nar/gkaa840
Open Targets Genetics: systematic identification of trait-associated genes using large-scale genetics and functional genomics
Abstract
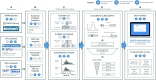
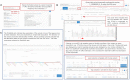
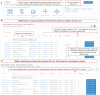
Open Targets Genetics (https://genetics.opentargets.org) is an open-access integrative resource that aggregates human GWAS and functional genomics data including gene expression, protein abundance, chromatin interaction and conformation data from a wide range of cell types and tissues to make robust connections between GWAS-associated loci, variants and likely causal genes. This enables systematic identification and prioritisation of likely causal variants and genes across all published trait-associated loci. In this paper, we describe the public resources we aggregate, the technology and analyses we use, and the functionality that the portal offers. Open Targets Genetics can be searched by variant, gene or study/phenotype. It offers tools that enable users to prioritise causal variants and genes at disease-associated loci and access systematic cross-disease and disease-molecular trait colocalization analysis across 92 cell types and tissues including the eQTL Catalogue. Data visualizations such as Manhattan-like plots, regional plots, credible sets overlap between studies and PheWAS plots enable users to explore GWAS signals in depth. The integrated data is made available through the web portal, for bulk download and via a GraphQL API, and the software is open source. Applications of this integrated data include identification of novel targets for drug discovery and drug repurposing.
© The Author(s) 2020. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures


References
-
- Hay M., Thomas D.W., Craighead J.L., Economides C., Rosenthal J.. Clinical development success rates for investigational drugs. Nat. Biotechnol. 2014; 32:40–51. - PubMed
-
- DiMasi J.A., Grabowski H.G., Hansen R.W.. Innovation in the pharmaceutical industry: new estimates of R&D costs. J. Health Econ. 2016; 47:20–33. - PubMed
-
- Nelson M.R, Tipney H., Painter J.L., Shen J., Nicoletti P., Shen Y., Floratos A., Sham P.C., Li M.J., Wang J. et al. .. The support of human genetic evidence for approved drug indications. Nat. Genet. 2019; 47:856–860. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

