A Cyclin D1-Dependent Transcriptional Program Predicts Clinical Outcome in Mantle Cell Lymphoma
- PMID: 33046520
- PMCID: PMC8051616
- DOI: 10.1158/1078-0432.CCR-20-2868
A Cyclin D1-Dependent Transcriptional Program Predicts Clinical Outcome in Mantle Cell Lymphoma
Abstract
Purpose: Mantle cell lymphoma (MCL) is characterized by the t(11;14)(q13;q32) translocation leading to cyclin D1 overexpression. Cyclin D1 is a major cell-cycle regulator and also regulates transcription, but the impact of cyclin D1-mediated transcriptional dysregulation on MCL pathogenesis remains poorly understood. The aim of this study was to define a cyclin D1-dependent gene expression program and analyze its prognostic value.
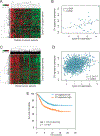
Experimental design: We integrated genome-wide expression analysis of cyclin D1-silenced and overexpressing cells with cyclin D1 chromatin-binding profiles to identify a cyclin D1-dependent transcriptional program in MCL cells. We analyzed this gene program in two MCL series of peripheral blood samples (n = 53) and lymphoid tissues (n = 106) to determine its biological and clinical relevance. We then obtained a simplified signature of this program and evaluated a third series of peripheral blood MCL samples (n = 81) by NanoString gene expression profiling to validate our findings.
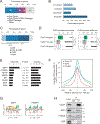
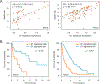
Results: We identified a cyclin D1-dependent transcriptional program composed of 295 genes that were mainly involved in cell-cycle control. The cyclin D1-dependent gene program was overexpressed in MCL tumors directly proportional to cyclin D1 levels. High expression of this program conferred an adverse prognosis with significant shorter overall survival of the patients. These observations were validated in an independent cohort of patients using a simplified 37-gene cyclin D1 signature. The cyclin D1-dependent transcriptional program was also present in multiple myeloma and breast tumors with cyclin D1 overexpression.
Conclusions: We identified a cyclin D1-dependent transcriptional program that is overexpressed in MCL and predicts clinical outcome.
©2020 American Association for Cancer Research.
Figures



References
-
- Bosch F, Jares P, Campo E, Lopez-Guillermo A, Piris MA, Villamor N, et al. PRAD-1/cyclin D1 gene overexpression in chronic lymphoproliferative disorders: a highly specific marker of mantle cell lymphoma. Blood. 1994;84:2726–32. - PubMed
-
- Seto M, Yamamoto K, Iida S, Akao Y, Utsumi KR, Kubonishi I, et al. Gene rearrangement and overexpression of PRAD1 in lymphoid malignancy with t(11;14)(q13;q32) translocation. Oncogene. 1992;7:1401–6. - PubMed
-
- Rosenwald A, Wright G, Wiestner A, Chan WC, Connors JM, Campo E, et al. The proliferation gene expression signature is a quantitative integrator of oncogenic events that predicts survival in mantle cell lymphoma. Cancer Cell. 2003;3:185–97. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

