Protein ontology on the semantic web for knowledge discovery
- PMID: 33046717
- PMCID: PMC7550340
- DOI: 10.1038/s41597-020-00679-9
Protein ontology on the semantic web for knowledge discovery
Abstract
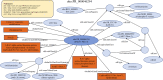
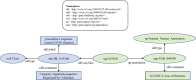
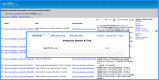
The Protein Ontology (PRO) provides an ontological representation of protein-related entities, ranging from protein families to proteoforms to complexes. Protein Ontology Linked Open Data (LOD) exposes, shares, and connects knowledge about protein-related entities on the Semantic Web using Resource Description Framework (RDF), thus enabling integration with other Linked Open Data for biological knowledge discovery. For example, proteins (or variants thereof) can be retrieved on the basis of specific disease associations. As a community resource, we strive to follow the Findability, Accessibility, Interoperability, and Reusability (FAIR) principles, disseminate regular updates of our data, support multiple methods for accessing, querying and downloading data in various formats, and provide documentation both for scientists and programmers. PRO Linked Open Data can be browsed via faceted browser interface and queried using SPARQL via YASGUI. RDF data dumps are also available for download. Additionally, we developed RESTful APIs to support programmatic data access. We also provide W3C HCLS specification compliant metadata description for our data. The PRO Linked Open Data is available at https://lod.proconsortium.org/ .
Conflict of interest statement
The authors declare no competing interests.
Figures



References
-
- Berners-Lee, T. Linked Data, https://www.w3.org/DesignIssues/LinkedData.html (2006).
-
- Callahan, A. et al. Bio2RDF release 2: improved coverage, interoperability and provenance of life science linked data. In: Cimiano P., Corcho O., Presutti V., Hollink L., Rudolph S. (eds) The Semantic Web: Semantics and Big Data. ESWC 2013. Lecture Notes in Computer Science. 7882, 200-212 (Springer, Berlin, Heidelberg, 2013).
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

