Transcriptional characterization of conjunctival melanoma identifies the cellular tumor microenvironment and prognostic gene signatures
- PMID: 33046735
- PMCID: PMC7550331
- DOI: 10.1038/s41598-020-72864-0
Transcriptional characterization of conjunctival melanoma identifies the cellular tumor microenvironment and prognostic gene signatures
Abstract
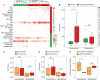
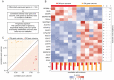
This study characterizes the transcriptome and the cellular tumor microenvironment (TME) of conjunctival melanoma (CM) and identifies prognostically relevant biomarkers. 12 formalin-fixed and paraffin-embedded CM were analyzed by MACE RNA sequencing, including six cases each with good or poor clinical outcome, the latter being defined by local recurrence and/or systemic metastases. Eight healthy conjunctival specimens served as controls. The TME of CM, as determined by bioinformatic cell type enrichment analysis, was characterized by the enrichment of melanocytes, pericytes and especially various immune cell types, such as plasmacytoid dendritic cells, natural killer T cells, B cells and mast cells. Differentially expressed genes between CM and control were mainly involved in inhibition of apoptosis, proteolysis and response to growth factors. POU3F3, BIRC5 and 7 were among the top expressed genes associated with inhibition of apoptosis. 20 genes, among them CENPK, INHA, USP33, CASP3, SNORA73B, AAR2, SNRNP48 and GPN1, were identified as prognostically relevant factors reaching high classification accuracy (area under the curve: 1.0). The present study provides new insights into the TME and the transcriptional profile of CM and additionally identifies new prognostic biomarkers. These results add new diagnostic tools and may lead to new options of targeted therapy for CM.
Conflict of interest statement
The authors declare no competing interests.
Figures



References
-
- Yu GP, Hu DN, McCormick S, Finger PT. Conjunctival melanoma: is it increasing in the United States? Am. J. Ophthalmol. 2003;135:800–806. - PubMed
-
- McLaughlin CC, et al. Incidence of noncutaneous melanomas in the U.S. Cancer. 2005;103:1000–1007. - PubMed
-
- Atzrodt L, Lapp T, Reinhard T, Auw-Haedrich C. Transformation of a naevus into a mixed pyogenic granuloma-naevus mimicking conjunctival melanoma. Pathology. 2020;52:269–271. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

