Mitochondrial genome sequencing and phylogeny of Haemagogus albomaculatus, Haemagogus leucocelaenus, Haemagogus spegazzinii, and Haemagogus tropicalis (Diptera: Culicidae)
- PMID: 33046768
- PMCID: PMC7550346
- DOI: 10.1038/s41598-020-73790-x
Mitochondrial genome sequencing and phylogeny of Haemagogus albomaculatus, Haemagogus leucocelaenus, Haemagogus spegazzinii, and Haemagogus tropicalis (Diptera: Culicidae)
Abstract
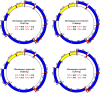
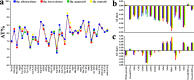
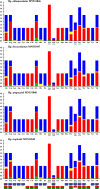
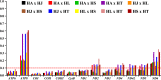
The genus Haemagogus (Diptera: Culicidae) comprises species of great epidemiological relevance, involved in transmission cycles of the Yellow fever virus and other arboviruses in South America. So far, only Haemagogus janthinomys has complete mitochondrial sequences available. Given the unavailability of information related to aspects of the evolutionary biology and molecular taxonomy of this genus, we report here, the first sequencing of the mitogenomes of Haemagogus albomaculatus, Haemagogus leucocelaenus, Haemagogus spegazzinii, and Haemagogus tropicalis. The mitogenomes showed an average length of 15,038 bp, average AT content of 79.3%, positive AT-skews, negative GC-skews, and comprised 37 functional subunits (13 PCGs, 22 tRNA, and 02 rRNA). The PCGs showed ATN as start codon, TAA as stop codon, and signs of purifying selection. The tRNAs had the typical leaf clover structure, except tRNASer1. Phylogenetic analyzes of Bayesian inference and Maximum Likelihood, based on concatenated sequences from all 13 PCGs, produced identical topologies and strongly supported the monophyletic relationship between the Haemagogus and Conopostegus subgenera, and corroborated with the known taxonomic classification of the evaluated taxa, based on external morphological aspects. The information produced on the mitogenomes of the Haemagogus species evaluated here may be useful in carrying out future taxonomic and evolutionary studies of the genus.
Conflict of interest statement
The authors declare no competing interests.
Figures

Similar articles
-
Description of mitochon genome and phylogenetic considerations of Sabethes bipartipes, Sabethes cyaneus, Sabethes quasicyaneus, and Sabethes tarsopus (Diptera: Culicidae).Acta Trop. 2022 Aug;232:106493. doi: 10.1016/j.actatropica.2022.106493. Epub 2022 May 4. Acta Trop. 2022. PMID: 35525314
-
First Description of the Mitogenome and Phylogeny of Culicinae Species from the Amazon Region.Genes (Basel). 2021 Dec 14;12(12):1983. doi: 10.3390/genes12121983. Genes (Basel). 2021. PMID: 34946932 Free PMC article.
-
Characterization of mitochondrial genome of Haemagogus janthinomys (Diptera: Culicidae).Mitochondrial DNA A DNA Mapp Seq Anal. 2017 Jan;28(1):50-51. doi: 10.3109/19401736.2015.1110793. Epub 2015 Dec 28. Mitochondrial DNA A DNA Mapp Seq Anal. 2017. PMID: 26709451
-
COI DNA barcoding to differentiate Haemagogus janthinomys and Haemagogus capricornii (Diptera: Culicidae) mosquitoes.Acta Trop. 2024 Nov;259:107377. doi: 10.1016/j.actatropica.2024.107377. Epub 2024 Sep 6. Acta Trop. 2024. PMID: 39245155
-
The complete mitochondrial genome of Anopheles minimus (Diptera: Culicidae) and the phylogenetics of known Anopheles mitogenomes.Insect Sci. 2016 Jun;23(3):353-65. doi: 10.1111/1744-7917.12326. Epub 2016 May 5. Insect Sci. 2016. PMID: 26852698
Cited by
-
Sequencing and Description of the Mitochondrial Genome of Orthopodomyia fascipes (Diptera: Culicidae).Genes (Basel). 2024 Jul 3;15(7):874. doi: 10.3390/genes15070874. Genes (Basel). 2024. PMID: 39062653 Free PMC article.
-
Phylogenetic Analysis of Some Species of the Anopheles hyrcanus Group (Diptera: Culicidae) in China Based on Complete Mitochondrial Genomes.Genes (Basel). 2023 Jul 16;14(7):1453. doi: 10.3390/genes14071453. Genes (Basel). 2023. PMID: 37510357 Free PMC article.
-
A mitochondrial genome phylogeny of voles and lemmings (Rodentia: Arvicolinae): Evolutionary and taxonomic implications.PLoS One. 2021 Nov 19;16(11):e0248198. doi: 10.1371/journal.pone.0248198. eCollection 2021. PLoS One. 2021. PMID: 34797834 Free PMC article.
-
Mitogenomes of mosquito species of Harris County in Texas.Sci Rep. 2025 Jul 1;15(1):22013. doi: 10.1038/s41598-025-04864-x. Sci Rep. 2025. PMID: 40593971 Free PMC article.
-
Mitochondrial Genome Variations and Possible Adaptive Implications in Some Tephritid Flies (Diptera, Tephritidae).Int J Mol Sci. 2025 Jun 10;26(12):5560. doi: 10.3390/ijms26125560. Int J Mol Sci. 2025. PMID: 40565023 Free PMC article.
References
-
- Arnell JH. Mosquito studies (Diptera, Culicidae). XXXII. A revision of the genus Haemagogus. Contrib. Am. Entomol. Inst. 1973;10:1–174.
-
- Forattini, O. P. Culicidologia Médica. Identificação, Biologia, Epidemiologia. Vol 2. (edusp, 2002).
-
- Zavortink TJ. Mosquito studies (Diptera, Culicidae). XXVIII. The new world species formerly placed in Aedes (Finlaya) Contrib. Am. Entomol. Inst. 1972;8:1–206.
-
- Marcondes C, Alencar J. Revisão de mosquitos Haemagogus Williston (Diptera: Culicidae) do Brasil. Rev Biomed. 2010;21:221–238.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

