Association between HLA gene polymorphisms and mortality of COVID-19: An in silico analysis
- PMID: 33047883
- PMCID: PMC7654404
- DOI: 10.1002/iid3.358
Association between HLA gene polymorphisms and mortality of COVID-19: An in silico analysis
Abstract
Introduction: The emergence of SARS-CoV-2 has caused global public health and economic crisis. Human leukocyte antigen (HLA) is a critical component of the viral antigen presentation pathway and plays essential roles in conferring differential viral susceptibility and severity of diseases. However, the association between HLA gene polymorphisms and risk for COVID-19 has not been fully elucidated. We hypothesized that HLA genotypes might impact on the differences in morbidity and mortality of COVID-19 across countries.
Methods: We conducted in silico analyses and examined an association of HLA gene polymorphisms with prevalence and mortality of COVID-19 by using publicly available databases.
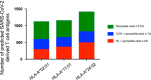
Results: We found that a possible association between HLA-A*02:01 and an increased risk for COVID-19. HLA-A*02:01 had a relatively lower capacity to present SARS-CoV-2 antigens compared with other frequent HLA class I molecules, HLA-A*11:01 or HLA-A*24:02.
Conclusion: This study suggests that individuals with HLA-A*11:01 or HLA-A*24:02 genotypes may generate efficiently T-cell-mediated antiviral responses to SARS-CoV-2 compared with HLA-A*02:01. The differences in HLA genotypes may potentially alter the course of the disease and its transmission.
Keywords: COVID-19; T cell; human leukocyte antigen; pandemic; severe acute respiratory syndrome coronavirus 2.
© 2020 The Authors. Immunity, Inflammation and Disease published by John Wiley & Sons Ltd.
Conflict of interest statement
The authors declare that there are no conflict of interests.
Figures


References
-
- Munster VJ, Koopmans M, van Doremalen N, van Riel D, de Wit E. A novel coronavirus emerging in China—key questions for impact assessment. N Engl J Med. 2020;382:692‐694. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

