Novel pathogenic genomic variants leading to autosomal dominant and recessive Robinow syndrome
- PMID: 33048444
- PMCID: PMC8445516
- DOI: 10.1002/ajmg.a.61908
Novel pathogenic genomic variants leading to autosomal dominant and recessive Robinow syndrome
Abstract
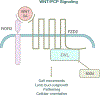
Robinow syndrome (RS) is a genetically heterogeneous disorder characterized by skeletal dysplasia and a distinctive facial appearance. Previous studies have revealed locus heterogeneity with rare variants in DVL1, DVL3, FZD2, NXN, ROR2, and WNT5A underlying the etiology of RS. The aforementioned "Robinow-associated genes" and their gene products all play a role in the WNT/planar cell polarity signaling pathway. We performed gene-targeted Sanger sequencing, exome sequencing, genome sequencing, and array comparative genomic hybridization on four subjects with a clinical diagnosis of RS who had not had prior DNA testing. Individuals in our cohort were found to carry pathogenic or likely pathogenic variants in three RS related genes: DVL1, ROR2, and NXN. One subject was found to have a nonsense variant (c.817C > T [p.Gln273*]) in NXN in trans with an ~1 Mb telomeric deletion on chromosome 17p containing NXN, which supports our contention that biallelic NXN variant alleles are responsible for a novel autosomal recessive RS locus. These findings provide increased understanding of the role of WNT signaling in skeletal development and maintenance. These data further support the hypothesis that dysregulation of the noncanonical WNT pathway in humans gives rise to RS.
Keywords: clinical diagnosis; deletion; missense; skeletal dysplasia; structural variant.
© 2020 Wiley Periodicals LLC.
Conflict of interest statement
Disclosures
Baylor College of Medicine (BCM) and Miraca Holdings have formed a joint venture with shared ownership and governance of the Baylor Genetics (BG), which performs clinical microarray analysis and clinical exome sequencing. J.R.L. serves on the Scientific Advisory Board of the BG. J.R.L. has stock ownership in 23andMe, is a paid consultant for Regeneron Pharmaceuticals, and is a coinventor on multiple United States and European patents related to molecular diagnostics for inherited neuropathies, eye diseases, and bacterial genomic fingerprinting. The Department of Molecular and Human Genetics at Baylor College of Medicine derives revenue from molecular genetic and clinical genomics testing offered at BG. The other authors declare no competing financial interests.
Figures

References
-
- Ali BR, Jeffery S, Patel N, Tinworth LE, Meguid N, Patton MA, & Afzal AR (2007). Novel Robinow syndrome causing mutations in the proximal region of the frizzled-like domain of ROR2 are retained in the endoplasmic reticulum. Human Genetics, 122(3–4), 389–395. doi:10.1007/s00439-007-0409-0 - DOI - PubMed
Publication types
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

