Age-induced mitochondrial DNA point mutations are inadequate to alter metabolic homeostasis in response to nutrient challenge
- PMID: 33049094
- PMCID: PMC7681042
- DOI: 10.1111/acel.13166
Age-induced mitochondrial DNA point mutations are inadequate to alter metabolic homeostasis in response to nutrient challenge
Abstract
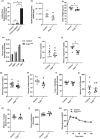
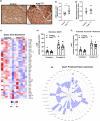
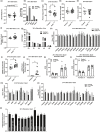
Mitochondrial dysfunction is frequently associated with impairment in metabolic homeostasis and insulin action, and is thought to underlie cellular aging. However, it is unclear whether mitochondrial dysfunction is a cause or consequence of insulin resistance in humans. To determine the impact of intrinsic mitochondrial dysfunction on metabolism and insulin action, we performed comprehensive metabolic phenotyping of the polymerase gamma (PolG) D257A "mutator" mouse, a model known to accumulate supraphysiological mitochondrial DNA (mtDNA) point mutations. We utilized the heterozygous PolG mutator mouse (PolG+/mut ) because it accumulates mtDNA point mutations ~ 500-fold > wild-type mice (WT), but fails to develop an overt progeria phenotype, unlike PolGmut/mut animals. To determine whether mtDNA point mutations induce metabolic dysfunction, we examined male PolG+/mut mice at 6 and 12 months of age during normal chow feeding, after 24-hr starvation, and following high-fat diet (HFD) feeding. No marked differences were observed in glucose homeostasis, adiposity, protein/gene markers of metabolism, or oxygen consumption in muscle between WT and PolG+/mut mice during any of the conditions or ages studied. However, proteomic analyses performed on isolated mitochondria from 12-month-old PolG+/mut mouse muscle revealed alterations in the expression of mitochondrial ribosomal proteins, electron transport chain components, and oxidative stress-related factors compared with WT. These findings suggest that mtDNA point mutations at levels observed in mammalian aging are insufficient to disrupt metabolic homeostasis and insulin action in male mice.
Keywords: POLG; aging; insulin resistance; metabolism; mitochondria; mitochondrial DNA; obesity.
© 2020 The Authors. Aging Cell published by the Anatomical Society and John Wiley & Sons Ltd.
Conflict of interest statement
The authors declare no competing financial interests.
Figures



References
-
- Ahlqvist, K. J. , Leoncini, S. , Pecorelli, A. , Wortmann, S. B. , Ahola, S. , Forsström, S. , … Suomalainen, A. (2015). MtDNA mutagenesis impairs elimination of mitochondria during erythroid maturation leading to enhanced erythrocyte destruction. Nature Communications, 6, 6494 10.1038/ncomms7494 - DOI - PubMed
-
- Avital, G. , Buchshtav, M. , Zhidkov, I. , Tuval (Feder), J. , Dadon, S. , Rubin, E. , … Mishmar, D. (2012). Mitochondrial DNA heteroplasmy in diabetes and normal adults: Role of acquired and inherited mutational patterns in twins. Human Molecular Genetics, 21(19), 4214–4224. 10.1093/hmg/dds245 - DOI - PMC - PubMed
-
- Center, M.A.M.D. Specializing in Mitochondrial Disease Treatment and Research. University of California, San Diego.
-
- Chella Krishnan, K. , Kurt, Z. , Barrere‐Cain, R. , Sabir, S. , Das, A. , Floyd, R. , … Yang, X. (2018). Integration of multi‐omics data from mouse diversity panel highlights mitochondrial dysfunction in non‐alcoholic fatty liver disease. Cell Systems, 6(1), 103–115 e107 10.1016/j.cels.2017.12.006 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- T32 HD007228/HD/NICHD NIH HHS/United States
- T32 AR065972/AR/NIAMS NIH HHS/United States
- T32 GM065823/GM/NIGMS NIH HHS/United States
- U54 DK120342/DK/NIDDK NIH HHS/United States
- P30 AG028748/AG/NIA NIH HHS/United States
- UL1 TR001881/TR/NCATS NIH HHS/United States
- UL1 TR000124/TR/NCATS NIH HHS/United States
- F31 DK108657/DK/NIDDK NIH HHS/United States
- P30 DK063491/DK/NIDDK NIH HHS/United States
- R01 AG055518/AG/NIA NIH HHS/United States
- T32 GM008243/GM/NIGMS NIH HHS/United States
- U54 AR052646/AR/NIAMS NIH HHS/United States
- K02 AG059847/AG/NIA NIH HHS/United States
- U24 DK097748/DK/NIDDK NIH HHS/United States
- R01 DK109724/DK/NIDDK NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources

