Challenges of Using Expansion Microscopy for Super-resolved Imaging of Cellular Organelles
- PMID: 33049107
- PMCID: PMC7894168
- DOI: 10.1002/cbic.202000571
Challenges of Using Expansion Microscopy for Super-resolved Imaging of Cellular Organelles
Abstract
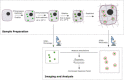
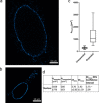
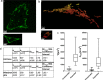
Expansion microscopy (ExM) has been successfully used to improve the spatial resolution when imaging tissues by optical microscopy. In ExM, proteins of a fixed sample are crosslinked to a swellable acrylamide gel, which expands when incubated in water. Therefore, ExM allows enlarged subcellular structures to be resolved that would otherwise be hidden to standard confocal microscopy. Herein, we aim to validate ExM for the study of peroxisomes, mitochondria, nuclei and the plasma membrane. Upon comparison of the expansion factors of these cellular compartments in HEK293 cells within the same gel, we found significant differences, of a factor of above 2, in expansion factors. For peroxisomes, the expansion factor differed even between peroxisomal membrane and matrix marker; this underlines the need for a thorough validation of expansion factors of this powerful technique. We further give an overview of possible quantification methods for the determination of expansion factors of intracellular organelles, and we highlight some potentials and challenges.
Keywords: STED microscopy; bioorganic chemistry; cell organelles; expansion microscopy; peptides.
© 2020 The Authors. ChemBioChem published by Wiley-VCH GmbH.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
-
- Gao M., et al., ACS Nano 2018, 12, 4178–4185. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

