Jasmonic Acid at the Crossroads of Plant Immunity and Pseudomonas syringae Virulence
- PMID: 33050569
- PMCID: PMC7589129
- DOI: 10.3390/ijms21207482
Jasmonic Acid at the Crossroads of Plant Immunity and Pseudomonas syringae Virulence
Abstract
Sensing of pathogen infection by plants elicits early signals that are transduced to affect defense mechanisms, such as effective blockage of pathogen entry by regulation of stomatal closure, cuticle, or callose deposition, change in water potential, and resource acquisition among many others. Pathogens, on the other hand, interfere with plant physiology and protein functioning to counteract plant defense responses. In plants, hormonal homeostasis and signaling are tightly regulated; thus, the phytohormones are qualified as a major group of signaling molecules controlling the most widely tinkered regulatory networks of defense and counter-defense strategies. Notably, the phytohormone jasmonic acid mediates plant defense responses to a wide array of pathogens. In this review, we present the synopsis on the jasmonic acid metabolism and signaling, and the regulatory roles of this hormone in plant defense against the hemibiotrophic bacterial pathogen Pseudomonas syringae. We also elaborate on how this pathogen releases virulence factors and effectors to gain control over plant jasmonic acid signaling to effectively cause disease. The findings discussed in this review may lead to ideas for the development of crop cultivars with enhanced disease resistance by genetic manipulation.
Keywords: Pseudomonas syringae; effectors; jasmonates; salicylic acid; stomata.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
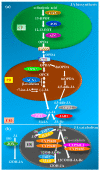

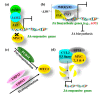

References
-
- Thomma B.P.H.J., Eggermont K., Penninckx I.A.M.A., Mauch-Mani B., Vogelsang R., Cammue B.P.A., Broekaert W.F. Separate jasmonate-dependent and salicylate-dependent defense-response pathways in Arabidopsis are essential for resistance to distinct microbial pathogens. Proc. Natl. Acad. Sci. USA. 1998;95:15107–15111. doi: 10.1073/pnas.95.25.15107. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

