Co-crystal structures of HIV TAR RNA bound to lab-evolved proteins show key roles for arginine relevant to the design of cyclic peptide TAR inhibitors
- PMID: 33051202
- PMCID: PMC7864049
- DOI: 10.1074/jbc.RA120.015444
Co-crystal structures of HIV TAR RNA bound to lab-evolved proteins show key roles for arginine relevant to the design of cyclic peptide TAR inhibitors
Abstract
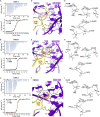
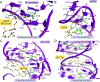
RNA-protein interfaces control key replication events during the HIV-1 life cycle. The viral trans-activator of transcription (Tat) protein uses an archetypal arginine-rich motif (ARM) to recruit the host positive transcription elongation factor b (pTEFb) complex onto the viral trans-activation response (TAR) RNA, leading to activation of HIV transcription. Efforts to block this interaction have stimulated production of biologics designed to disrupt this essential RNA-protein interface. Here, we present four co-crystal structures of lab-evolved TAR-binding proteins (TBPs) in complex with HIV-1 TAR. Our results reveal that high-affinity binding requires a distinct sequence and spacing of arginines within a specific β2-β3 hairpin loop that arose during selection. Although loops with as many as five arginines were analyzed, only three arginines could bind simultaneously with major-groove guanines. Amino acids that promote backbone interactions within the β2-β3 loop were also observed to be important for high-affinity interactions. Based on structural and affinity analyses, we designed two cyclic peptide mimics of the TAR-binding β2-β3 loop sequences present in two high-affinity TBPs (KD values of 4.2 ± 0.3 and 3.0 ± 0.3 nm). Our efforts yielded low-molecular weight compounds that bind TAR with low micromolar affinity (KD values ranging from 3.6 to 22 μm). Significantly, one cyclic compound within this series blocked binding of the Tat-ARM peptide to TAR in solution assays, whereas its linear counterpart did not. Overall, this work provides insight into protein-mediated TAR recognition and lays the ground for the development of cyclic peptide inhibitors of a vital HIV-1 RNA-protein interaction.
Keywords: HIV TAR; HIV Tat; RNA structure; RNA-binding protein; RNA-protein interaction; RNA-protein interactions; X-ray crystallography; arginine-rich domain; cyclic peptide; cyclic peptide inhibitor; drug discovery; human immunodeficiency virus (HIV); isothermal titration calorimetry; isothermal titration calorimetry (ITC); peptide chemical synthesis; surface plasmon resonance; surface plasmon resonance (SPR).
© 2020 Chavali et al.
Conflict of interest statement
Conflict of interest—The authors declare that they have no conflicts of interest with the contents of this article.
Figures





References
Uncited reference
-
- Hodel A., Kim S.H., Brunger A.T. Model bias in macromolecular crystal-structures. Acta Crystallogr. A. 1992;48:851–858. doi: 10.1107/S0108767392006044. - DOI
-
- Brunger A.T., Adams P.D., Clore G.M., DeLano W.L., Gros P., Grosse-Kunstleve R.W., Jiang J.S., Kuszewski J., Nilges M., Pannu N.S., Read R.J., Rice L.M., Simonson T., Warren G.L. Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. 9757107. - DOI - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

