Blood functional assay for rapid clinical interpretation of germline TP53 variants
- PMID: 33051313
- PMCID: PMC8639931
- DOI: 10.1136/jmedgenet-2020-107059
Blood functional assay for rapid clinical interpretation of germline TP53 variants
Abstract
Background: The interpretation of germline TP53 variants is critical to ensure appropriate medical management of patients with cancer and follow-up of variant carriers. This interpretation remains complex and is becoming a growing challenge considering the exponential increase in TP53 tests. We developed a functional assay directly performed on patients' blood.
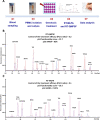
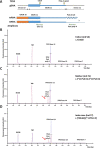
Methods: Peripheral blood mononuclear cells were cultured, activated, exposed to doxorubicin and the p53-mediated transcriptional response was quantified using reverse transcription-multiplex ligation probe amplification and RT-QMPSF assays, including 10 p53 targets selected from transcriptome analysis, and two amplicons to measure p53 mRNA levels. We applied this blood functional assay to 77 patients addressed for TP53 analysis.
Results: In 51 wild-type TP53 individuals, the mean p53 functionality score was 12.7 (range 7.5-22.8). Among eight individuals harbouring likely pathogenic or pathogenic variants, the scores were reduced (mean 4.8, range 3.1-7.1), and p53 mRNA levels were reduced in patients harbouring truncating variants. We tested 14 rare unclassified variants (p.(Pro72His), p.(Gly105Asp), p.(Arg110His), p.(Phe134Leu), p.(Arg158Cys), p.(Pro191Arg), p.(Pro278Arg), p.(Arg283Cys), p.(Leu348Ser), p.(Asp352Tyr), p.(Gly108_Phe109delinsVal), p.(Asn131del), p.(Leu265del), c.-117G>T) and 12 yielded functionally abnormal scores. Remarkably, the assay revealed that the c.*1175A>C polymorphic variant within TP53 poly-adenylation site can impact p53 function with the same magnitude as a null variant, when present on both alleles, and may act as a modifying factor in pathogenic variant carriers.
Conclusion: This blood p53 assay should therefore be a useful tool for the rapid clinical classification of germline TP53 variants and detection of non-coding functional variants.
Keywords: clinical laboratory techniques; genetic predisposition to disease; genetic testing; germ-line mutation; methods.
© Author(s) (or their employer(s)) 2021. Re-use permitted under CC BY-NC. No commercial re-use. See rights and permissions. Published by BMJ.
Conflict of interest statement
Competing interests: None declared.
Figures

References
-
- Bougeard G, Renaux-Petel M, Flaman J-M, Charbonnier C, Fermey P, Belotti M, Gauthier-Villars M, Stoppa-Lyonnet D, Consolino E, Brugières L, Caron O, Benusiglio PR, Bressac-de Paillerets B, Bonadona V, Bonaïti-Pellié C, Tinat J, Baert-Desurmont S, Frebourg T. Revisiting Li-Fraumeni syndrome from TP53 mutation carriers. J Clin Oncol 2015;33:2345–52. 10.1200/JCO.2014.59.5728 - DOI - PubMed
-
- Kasper E, Angot E, Colasse E, Nicol L, Sabourin J-C, Adriouch S, Lacoume Y, Charbonnier C, Raad S, Frebourg T, Flaman J-M, Bougeard G. Contribution of genotoxic anticancer treatments to the development of multiple primary tumours in the context of germline TP53 mutations. Eur J Cancer 2018;101:254–62. 10.1016/j.ejca.2018.06.011 - DOI - PubMed
-
- Villani A, Shore A, Wasserman JD, Stephens D, Kim RH, Druker H, Gallinger B, Naumer A, Kohlmann W, Novokmet A, Tabori U, Tijerin M, Greer M-LC, Finlay JL, Schiffman JD, Malkin D. Biochemical and imaging surveillance in germline TP53 mutation carriers with Li-Fraumeni syndrome: 11 year follow-up of a prospective observational study. Lancet Oncol 2016;17:1295–305. 10.1016/S1470-2045(16)30249-2 - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous
