Anti-oncogene PTPN13 inactivation by hepatitis B virus X protein counteracts IGF2BP1 to promote hepatocellular carcinoma progression
- PMID: 33051595
- PMCID: PMC7790756
- DOI: 10.1038/s41388-020-01498-3
Anti-oncogene PTPN13 inactivation by hepatitis B virus X protein counteracts IGF2BP1 to promote hepatocellular carcinoma progression
Abstract
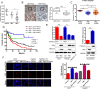
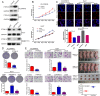
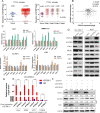
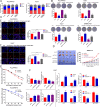
Hepatitis B x protein (HBx) affects cellular protein expression and participates in the tumorigenesis and progression of hepatitis B virus (HBV)-related hepatocellular carcinoma (HCC). Metabolic reprogramming contributed to the HCC development, but its role in HBV-related HCC remains largely unclear. Tyrosine-protein phosphatase nonreceptor type 13 (PTPN13) is a significant regulator in tumor development, however, its specific role in hepatocarcinogenesis remains to be explored. Here, we found that decreased PTPN13 expression was associated with HBV/HBx. Patients with low PTPN13 expression showed a poor prognosis. Functional assays revealed that PTPN13 inhibited proliferation and tumorigenesis in vitro and in vivo. Further mechanistic studies indicated that HBx inhibited PTPN13 expression by upregulating the expression of DNMT3A and interacting with DNMT3A. Furthermore, we found that DNMT3A bound to the PTPN13 promoter (-343 to -313 bp) in an epigenetically controlled manner associated with elevated DNA methylation and then inhibited PTPN13 transcription. In addition, we identified IGF2BP1 as a novel PTPN13-interacting gene and demonstrated that PTPN13 influences c-Myc expression by directly and competitively binding to IGF2BP1 to decrease the intracellular concentration of functional IGF2BP1. Overexpressing PTPN13 promoted c-Myc mRNA degradation independent of the protein tyrosine phosphatase (PTP) activity of PTPN13. Importantly, we discovered that the PTPN13-IGF2BP1-c-Myc axis was important for cancer cell growth through promoting metabolic reprogramming. We verified the significant negative correlations between PTPN13 expression and c-Myc, PSPH, and SLC7A1 expression in clinical HCC tissue samples. In summary, our findings demonstrate that PTPN13 is a novel regulator of HBV-related hepatocarcinogenesis and may play an important role in HCC. PTPN13 may serve as a prognostic marker and therapeutic target in HBV-related HCC patients.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures




References
-
- Chen W, Zheng R, Baade PD, Zhang S, Zeng H, Bray F, et al. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66:115–32. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

