A high-throughput CRISPR interference screen for dissecting functional regulators of GPCR/cAMP signaling
- PMID: 33052901
- PMCID: PMC7588078
- DOI: 10.1371/journal.pgen.1009103
A high-throughput CRISPR interference screen for dissecting functional regulators of GPCR/cAMP signaling
Abstract
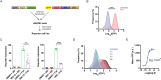
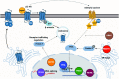
G protein-coupled receptors (GPCRs) allow cells to respond to chemical and sensory stimuli through generation of second messengers, such as cyclic AMP (cAMP), which in turn mediate a myriad of processes, including cell survival, proliferation, and differentiation. In order to gain deeper insights into the complex biology and physiology of these key cellular pathways, it is critical to be able to globally map the molecular factors that shape cascade function. Yet, to this date, efforts to systematically identify regulators of GPCR/cAMP signaling have been lacking. Here, we combined genome-wide screening based on CRISPR interference with a novel sortable transcriptional reporter that provides robust readout for cAMP signaling, and carried out a functional screen for regulators of the pathway. Due to the sortable nature of the platform, we were able to assay regulators with strong and moderate phenotypes by analyzing sgRNA distribution among three fractions with distinct reporter expression. We identified 45 regulators with strong and 50 regulators with moderate phenotypes not previously known to be involved in cAMP signaling. In follow-up experiments, we validated the functional effects of seven newly discovered mediators (NUP93, PRIM1, RUVBL1, PKMYT1, TP53, SF3A2, and HRAS), and showed that they control distinct steps of the pathway. Thus, our study provides proof of principle that the screening platform can be applied successfully to identify bona fide regulators of GPCR/second messenger cascades in an unbiased and high-throughput manner, and illuminates the remarkable functional diversity among GPCR regulators.
Conflict of interest statement
I have read the journal’s policy and the authors of this manuscript have the following competing interests: M.K. has filed a patent application related to CRISPRi and CRISPRa screening (PCT/US15/40449) and serves on the Scientific Advisory Board of Engine Biosciences.
Figures


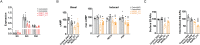
References
-
- Lohse MJ, Engelhardt S, Eschenhagen T. What is the role of beta-adrenergic signaling in heart failure? Circ Res. 2003;93(10):896–906. 10.1161/01.RES.0000102042.83024.CA . - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

