Analytical Sensitivity and Specificity of Two RT-qPCR Protocols for SARS-CoV-2 Detection Performed in an Automated Workflow
- PMID: 33053675
- PMCID: PMC7599812
- DOI: 10.3390/genes11101183
Analytical Sensitivity and Specificity of Two RT-qPCR Protocols for SARS-CoV-2 Detection Performed in an Automated Workflow
Abstract
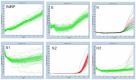
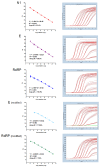
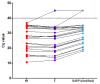
WHO declared the novel coronavirus (COVID-19) outbreak a global pandemic on 11 March 2020. The establishment of standardized RT-qPCR protocols for respiratory secretions testing, as well as sharing of specimens, data, and information became critical. Here, we investigate the analytical performance of two interim RT-qPCR protocols (Charité and Centers for Disease Control (CDC)) for the qualitative detection of SARS-CoV-2 executed in a fully automated platform. Analytical specificity, PCR amplification efficiency, analytical sensitivity (limit of detection), and cross-reactivity were evaluated using contrived samples. The on-going accuracy was evaluated by retrospective analysis of our test results database (real clinical samples). N1, E, and a modified version of RdRP assays presented adequate analytical specificity, amplification efficiency, and analytical sensitivity using contrived samples. The three assays were applied to all individuals who requested the SARS-CoV-2 molecular test assay in our laboratory and it was observed that N1 gave more positive results than E, and E gave more positive results than RdRP (modified). The RdRP and E were removed from the test and its final version, based on N1 assay only, was applied to 30,699 Brazilian individuals (from 19 February 2020 to 8 May 2020). The aggregated test results available in the database were also presented.
Keywords: RT-qPCR; SARS-CoV-2; validation.
Conflict of interest statement
Sabin laboratory is a private commercial laboratory. All authors are employees of Sabin Laboratory.
Figures



References
-
- World Health Organization World Experts and Funders Set Priorities for Covid-19 Research. [(accessed on 18 February 2020)]; Available online: https://http://www.who.int/news-room/detail/12-02-2020-world-experts-and....
-
- Corman V.M., Landt O., Kaiser M., Molenkamp R., Meijer A., Chu D.K., Bleicker T., Brunink S., Schneider J., Schmidt M.L., et al. Detection of 2019 novel coronavirus (2019-ncov) by real-time rt-pcr. Eurosurveill. Bull. Eur. Mal. Transmissibles Eur. Commun. Dis. Bull. 2020;25:2000045. doi: 10.2807/1560-7917.ES.2020.25.3.2000045. - DOI - PMC - PubMed
-
- World Health Organization Coronavirus Disease (Covid-19) Technical Guidance: Laboratory Testing for 2019-Ncov in Humans. [(accessed on 18 February 2020)]; Available online: https://http://www.who.int/emergencies/diseases/novel-coronavirus-2019/t....
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

