Characterization and evolutionary origin of novel C2H2 zinc finger protein (ZNF648) required for both erythroid and megakaryocyte differentiation in humans
- PMID: 33054117
- PMCID: PMC8561289
- DOI: 10.3324/haematol.2020.256347
Characterization and evolutionary origin of novel C2H2 zinc finger protein (ZNF648) required for both erythroid and megakaryocyte differentiation in humans
Abstract
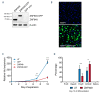
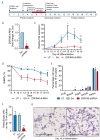
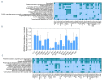
Human ZNF648 is a novel poly C-terminal C2H2 zinc finger protein identified amongst the most dysregulated proteins in erythroid cells differentiated from iPSC. Its nuclear localisation and structure indicate it is likely a DNA-binding protein. Using a combination of ZNF648 overexpression in an iPSC line and primary adult erythroid cells, ZNF648 knockdown in primary adult erythroid cells and megakaryocytes, comparative proteomics and transcriptomics we show that ZNF648 is required for both erythroid and megakaryocyte differentiation. Orthologues of ZNF648 were detected across Mammals, Reptilia, Actinopterygii, in some Aves, Amphibia and Coelacanthiformes suggesting the gene originated in the common ancestor of Osteichthyes (Euteleostomi or bony fish). Conservation of the C-terminal zinc finger domain is higher, with some variation in zinc finger number but a core of at least six zinc fingers conserved across all groups, with the N-terminus recognisably similar within but not between major lineages. This suggests the N-terminus of ZNF648 evolves faster than the C-terminus, however this is not due to exon-shuffling as the entire coding region of ZNF648 is within a single exon. As for other such transcription factors, the N-terminus likely carries out regulatory functions, but showed no sequence similarity to any known domains. The greater functional constraint on the zinc finger domain suggests ZNF648 binds at least some similar regions of DNA in the different organisms. However, divergence of the N-terminal region may enable differential expression, allowing adaptation of function in the different organisms.
Figures





References
-
- Klug A. The discovery of zinc fingers and their development for practical applications in gene regulation and genome manipulation. Q Rev Biophys. 2010;43(1):1-21. - PubMed
-
- Vaquerizas JM, Kummerfeld SK, Teichmann SA, et al. . A census of human transcription factors: function, expression and evolution. Nat Rev Genet. 2009;10(4):252-263. - PubMed
-
- Kim SI, Bresnick EH. Transcriptional control of erythropoiesis: emerging mechanisms and principles. Oncogene. 2007;26(47):6777-6794. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

