Genome-wide characterization and expression analysis of the heat shock transcription factor family in pumpkin (Cucurbita moschata)
- PMID: 33054710
- PMCID: PMC7557022
- DOI: 10.1186/s12870-020-02683-y
Genome-wide characterization and expression analysis of the heat shock transcription factor family in pumpkin (Cucurbita moschata)
Abstract
Background: Crop quality and yield are affected by abiotic and biotic stresses, and heat shock transcription factors (Hsfs) are considered to play important roles in regulating plant tolerance under various stresses. To investigate the response of Cucurbita moschata to abiotic stress, we analyzed the genome of C. moschata.
Results: In this research, a total of 36 C. moschata Hsf (CmHsf) members were identified and classified into three subfamilies (I, II, and III) according to their amino acid sequence identity. The Hsfs of the same subfamily usually exhibit a similar gene structure (intron-exon distribution) and conserved domains (DNA-binding and other functional domains). Chromosome localization analysis showed that the 36 CmHsfs were unevenly distributed on 18 of the 21 chromosomes (except for Cm_Chr00, Cm_Chr08 and Cm_Chr20), among which 18 genes formed 9 duplicated gene pairs that have undergone segmental duplication events. The Ka/Ks ratio showed that the duplicated CmHsfs have mainly experienced strong purifying selection. High-level synteny was observed between C. moschata and other Cucurbitaceae species.
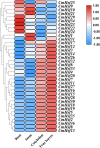
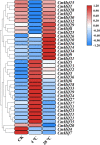
Conclusions: The expression profile of CmHsfs in the roots, stems, cotyledons and true leaves revealed that the CmHsfs exhibit tissue specificity. The analysis of cis-acting elements and quantitative real-time polymerase chain reaction (qRT-PCR) revealed that some key CmHsfs were activated by cold stress, heat stress, hormones and salicylic acid. This study lays the foundation for revealing the role of CmHsfs in resistance to various stresses, which is of great significance for the selection of stress-tolerant C. moschata.
Keywords: Cis-acting elements; Conserved domain; Cucurbita moschata; Expression pattern; Gene duplication; Heat shock transcription factor.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures







References
-
- Baniwal SK, Bharti K, Chan KY, Fauth M, Ganguli A, Kotak S, Mishra SK, Nover L, Port M, Scharf KD, Tripp J, Weber C, Zielinski D, Koskull DP. von. Heat stress response in plants: a complex game with chaperones and more than twenty heat stress transcription factors. J Biosci. 2004;29(4):471–487. doi: 10.1007/BF02712120. - DOI - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

