Circadian disruption promotes tumor-immune microenvironment remodeling favoring tumor cell proliferation
- PMID: 33055171
- PMCID: PMC7556830
- DOI: 10.1126/sciadv.aaz4530
Circadian disruption promotes tumor-immune microenvironment remodeling favoring tumor cell proliferation
Abstract
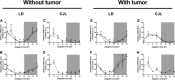
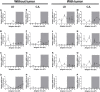
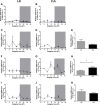
Circadian disruption negatively affects physiology, posing a global health threat that manifests in proliferative, metabolic, and immune diseases, among others. Because outputs of the circadian clock regulate daily fluctuations in the immune response, we determined whether circadian disruption results in tumor-associated immune cell remodeling, facilitating tumor growth. Our findings show that tumor growth rate increased and latency decreased under circadian disruption conditions compared to normal light-dark (LD) schedules in a murine melanoma model. Circadian disruption induced the loss or inversion of daily patterns of M1 (proinflammatory) and M2 (anti-inflammatory) macrophages and cytokine levels in spleen and tumor tissues. Circadian disruption also induced (i) deregulation of rhythmic expression of clock genes and (ii) of cyclin genes in the liver, (iii) increased CcnA2 levels in the tumor, and (iv) dampened expression of the cell cycle inhibitor p21WAF/CIP1 , all of which contribute to a proliferative phenotype.
Copyright © 2020 The Authors, some rights reserved; exclusive licensee American Association for the Advancement of Science. No claim to original U.S. Government Works. Distributed under a Creative Commons Attribution NonCommercial License 4.0 (CC BY-NC).
Figures



References
-
- Golombek D. A., Casiraghi L. P., Agostino P. V., Paladino N., Duhart J. M., Plano S. A., Chiesa J. J., The times they’re a-changing: Effects of circadian desynchronization on physiology and disease. J. Physiol. Paris 107, 310–322 (2013). - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

